You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001661_01280
You are here: Home > Sequence: MGYG000001661_01280
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides gallinarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides gallinarum | |||||||||||
| CAZyme ID | MGYG000001661_01280 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11457; End: 13964 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH43 | 32 | 319 | 2.9e-120 | 0.9928057553956835 |
| CE1 | 602 | 831 | 1.5e-56 | 0.9691629955947136 |
| CBM6 | 343 | 467 | 7.3e-35 | 0.8695652173913043 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd18618 | GH43_Xsa43E-like | 4.48e-150 | 39 | 321 | 1 | 274 | Glycosyl hydrolase family 43, including Butyrivibrio proteoclasticus arabinofuranosidase Xsa43E. This glycosyl hydrolase family 43 (GH43) subgroup belongs to the GH43_AXH-like subgroup which includes enzymes that have been characterized with beta-xylosidase (EC 3.2.1.37), alpha-L-arabinofuranosidase (EC 3.2.1.55), alpha-1,2-L-arabinofuranosidase 43A (arabinan-specific; EC 3.2.1.-), endo-alpha-L-arabinanase as well as arabinoxylan arabinofuranohydrolase (AXH) activities. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. AXHs specifically hydrolyze the glycosidic bond between arabinofuranosyl substituents and xylopyranosyl backbone residues of arabinoxylan. This subgroup includes Cellvibrio japonicus arabinan-specific alpha-1,2-arabinofuranosidase, CjAbf43A, which confers its specificity by a surface cleft that is complementary to the helical backbone of the polysaccharide, and Butyrivibrio proteoclasticus GH43 enzyme Xsa43E, also an arabinofuranosidase, which has been shown to cleave arabinose side chains from short segments of xylan. Several of these enzymes also contain carbohydrate binding modules (CBMs) that bind cellulose or xylan. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd08990 | GH43_AXH_like | 4.86e-109 | 42 | 322 | 2 | 269 | Glycosyl hydrolase family 43 protein, includes arabinoxylan arabinofuranohydrolase, beta-xylosidase, endo-1,4-beta-xylanase, and alpha-L-arabinofuranosidase. This subgroup includes Bacillus subtilis arabinoxylan arabinofuranohydrolase (XynD;BsAXH-m23;BSU18160), Butyrivibrio proteoclasticus alpha-L-arabinofuranosidase (Xsa43E;bpr_I2319), Clostridium stercorarium alpha-L-arabinofuranosidase XylA, and metagenomic beta-xylosidase (EC 3.2.1.37) / alpha-L-arabinofuranosidase (EC 3.2.1.55) CoXyl43. It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. The GH43_AXH-like subgroup includes enzymes that have been characterized with beta-xylosidase, alpha-L-arabinofuranosidase, endo-alpha-L-arabinanase as well as arabinoxylan arabinofuranohydrolase (AXH) activities. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. AXHs specifically hydrolyze the glycosidic bond between arabinofuranosyl substituents and xylopyranosyl backbone residues of arabinoxylan. Metagenomic beta-xylosidase/alpha-L-arabinofuranosidase CoXyl43 shows synergy with Trichoderma reesei cellulases and promotes plant biomass saccharification by degrading xylo-oligosaccharides, such as xylobiose and xylotriose, into the monosaccharide xylose. Studies show that the hydrolytic activity of CoXyl43 is stimulated in the presence of calcium. Several of these enzymes also contain carbohydrate binding modules (CBMs) that bind cellulose or xylan. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd09003 | GH43_XynD-like | 2.95e-70 | 32 | 323 | 1 | 315 | Glycosyl hydrolase family 43 protein such as Bacillus subtilis arabinoxylan arabinofuranohydrolase (XynD;BsAXH-m23;BSU18160). This glycosyl hydrolase family 43 (GH43) subgroup includes characterized Bacillus subtilis arabinoxylan arabinofuranohydrolase (AXH), Caldicellulosiruptor sp. Tok7B.1 beta-1,4-xylanase (EC 3.2.1.8) / alpha-L-arabinosidase (EC 3.2.1.55) XynA, Caldicellulosiruptor sp. Rt69B.1 xylanase C (EC 3.2.1.8) XynC, and Caldicellulosiruptor saccharolyticus beta-xylosidase (EC 3.2.1.37)/ alpha-L-arabinofuranosidase (EC 3.2.1.55) XynF. It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. It belongs to the GH43_AXH-like subgroup which includes enzymes that have been annotated as having beta-xylosidase, alpha-L-arabinofuranosidase and arabinoxylan alpha-L-1,3-arabinofuranohydrolase, xylanase (endo-alpha-L-arabinanase) as well as AXH activities. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. AXHs specifically hydrolyze the glycosidic bond between arabinofuranosyl substituents and xylopyranosyl backbone residues of arabinoxylan. Bacillus subtilis AXH (BsAXH-m2,3) has been shown to cleave arabinose units from O-2- or O-3-mono-substituted xylose residues and superposition of its structure with known structures of the GH43 exo-acting enzymes, beta-xylosidase and alpha-L-arabinanase, each in complex with their substrate, reveals a different orientation of the sugar backbone. Several of these enzymes also contain carbohydrate binding modules (CBMs) that bind cellulose or xylan. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd09004 | GH43_bXyl-like | 7.56e-70 | 42 | 320 | 2 | 263 | Glycosyl hydrolase family 43 protein such as Bacteroides thetaiotaomicron VPI-5482 alpha-L-arabinofuranosidases (BT3675;BT_3675) and (BT3662;BT_3662); includes mostly xylanases. This glycosyl hydrolase family 43 (GH43) subgroup includes enzymes that have been annotated as xylan-digesting beta-xylosidase (EC 3.2.1.37) and xylanase (endo-alpha-L-arabinanase, EC 3.2.1.8) activities, as well the Bacteroides thetaiotaomicron VPI-5482 alpha-L-arabinofuranosidases (EC 3.2.1.55) (BT3675;BT_3675) and (BT3662;BT_3662). It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd18619 | GH43_CoXyl43_like | 3.27e-58 | 33 | 320 | 1 | 311 | Glycosyl hydrolase family 43 protein such as metagenomic beta-xylosidase/alpha-L-arabinofuranosidase CoXyl43. This glycosyl hydrolase family 43 (GH43) subgroup belongs to the GH43_AXH-like subgroup which includes enzymes that have been characterized with beta-xylosidase (EC 3.2.1.37), alpha-L-arabinofuranosidase (EC 3.2.1.55), alpha-1,2-L-arabinofuranosidase 43A (arabinan-specific; EC 3.2.1.-), endo-alpha-L-arabinanase as well as arabinoxylan arabinofuranohydrolase (AXH) activities. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. Included in this subfamily is the metagenomic beta-xylosidase/alpha-L-arabinofuranosidase CoXyl43, which shows synergy with Trichoderma reesei cellulases and promotes plant biomass saccharification by degrading xylo-oligosaccharides, such as xylobiose and xylotriose, into the monosaccharide xylose. Studies show that the hydrolytic activity of CoXyl43 is stimulated in the presence of calcium. Several of these enzymes also contain carbohydrate binding modules (CBMs) that bind cellulose or xylan. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADD61495.1 | 0.0 | 46 | 835 | 1 | 790 |
| ALK86294.1 | 0.0 | 1 | 834 | 1 | 834 |
| ABR37762.1 | 0.0 | 1 | 834 | 1 | 834 |
| QQY40420.1 | 0.0 | 3 | 834 | 1 | 832 |
| QUT56982.1 | 0.0 | 3 | 834 | 1 | 832 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6MOU_A | 5.86e-115 | 473 | 834 | 21 | 380 | ChainA, Isoamylase N-terminal domain protein [Bacteroides intestinalis DSM 17393],6MOU_B Chain B, Isoamylase N-terminal domain protein [Bacteroides intestinalis DSM 17393] |
| 6MOT_A | 1.55e-114 | 474 | 834 | 1 | 359 | ChainA, Isoamylase N-terminal domain protein [Bacteroides intestinalis DSM 17393] |
| 7B5V_A | 7.27e-114 | 481 | 834 | 29 | 386 | ChainA, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48 [Dysgonomonas mossii DSM 22836],7B5V_B Chain B, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48 [Dysgonomonas mossii DSM 22836],7B6B_A Chain A, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48 [Dysgonomonas mossii DSM 22836],7B6B_B Chain B, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48 [Dysgonomonas mossii DSM 22836] |
| 4NOV_A | 1.41e-100 | 31 | 319 | 45 | 331 | Xsa43E,a GH43 family enzyme from Butyrivibrio proteoclasticus [Butyrivibrio proteoclasticus B316] |
| 6NE9_A | 3.42e-87 | 470 | 834 | 21 | 382 | ChainA, Isoamylase protein [Bacteroides intestinalis],6NE9_B Chain B, Isoamylase protein [Bacteroides intestinalis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EXZ4 | 7.39e-152 | 482 | 835 | 319 | 671 | Carbohydrate acetyl esterase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axe1-6A PE=1 SV=1 |
| D5EY13 | 2.61e-63 | 485 | 834 | 388 | 726 | Endo-1,4-beta-xylanase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyn10D-fae1A PE=1 SV=1 |
| Q45071 | 8.82e-62 | 28 | 465 | 36 | 510 | Arabinoxylan arabinofuranohydrolase OS=Bacillus subtilis (strain 168) OX=224308 GN=xynD PE=1 SV=2 |
| P45796 | 1.08e-60 | 32 | 514 | 39 | 565 | Arabinoxylan arabinofuranohydrolase OS=Paenibacillus polymyxa OX=1406 GN=xynD PE=1 SV=1 |
| P49943 | 2.30e-37 | 39 | 328 | 13 | 325 | Xylosidase/arabinosidase OS=Bacteroides ovatus OX=28116 GN=xsa PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
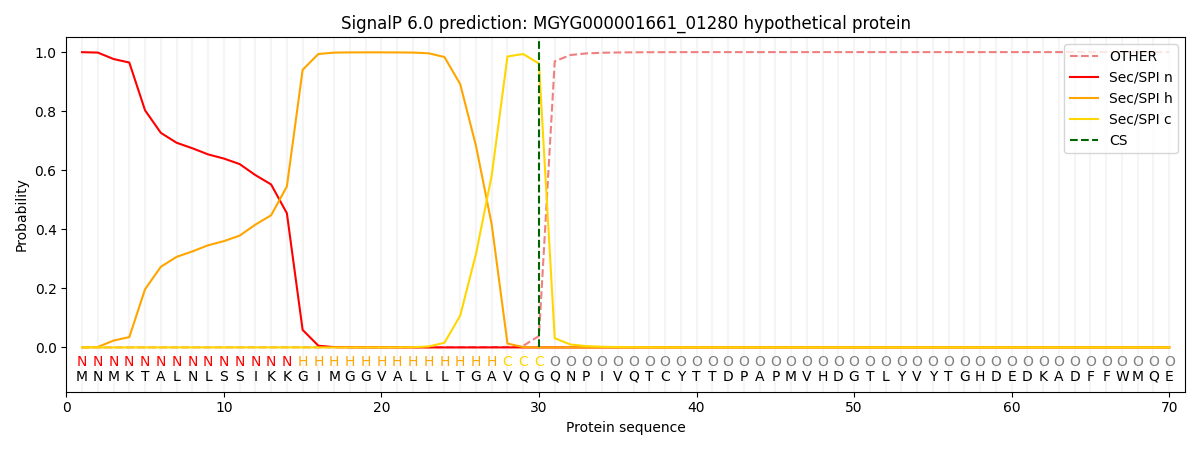
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000616 | 0.998210 | 0.000524 | 0.000249 | 0.000192 | 0.000169 |
