You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001661_02792
You are here: Home > Sequence: MGYG000001661_02792
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides gallinarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides gallinarum | |||||||||||
| CAZyme ID | MGYG000001661_02792 | |||||||||||
| CAZy Family | GH158 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20830; End: 22110 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH158 | 34 | 165 | 2.3e-32 | 0.9848484848484849 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.13e-06 | 23 | 138 | 286 | 410 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| COG3794 | PetE | 0.002 | 340 | 405 | 53 | 111 | Plastocyanin [Energy production and conversion]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SIP56416.1 | 2.67e-230 | 2 | 425 | 4 | 429 |
| SIP56393.1 | 2.67e-230 | 2 | 425 | 4 | 429 |
| SIP56346.1 | 2.67e-230 | 2 | 425 | 4 | 429 |
| SIP56337.1 | 2.67e-230 | 2 | 425 | 4 | 429 |
| QPH56445.1 | 2.10e-229 | 2 | 425 | 4 | 428 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PAL_A | 9.33e-229 | 22 | 425 | 3 | 406 | Bacteroidesuniformis endo-laminarinase BuGH158 from the beta(1,3)-glucan utilization locus [Bacteroides uniformis],6PAL_B Bacteroides uniformis endo-laminarinase BuGH158 from the beta(1,3)-glucan utilization locus [Bacteroides uniformis] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
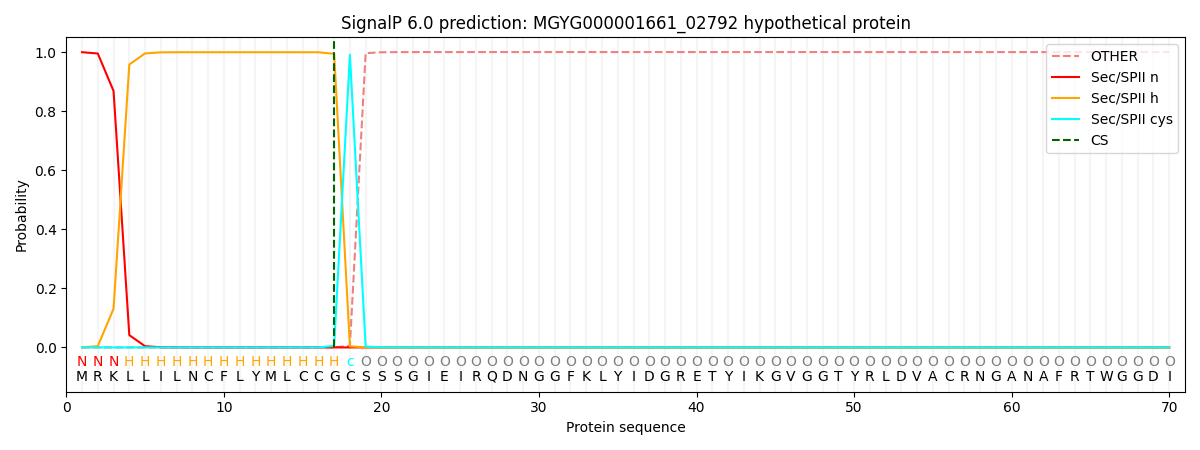
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000385 | 0.999677 | 0.000000 | 0.000000 | 0.000000 |
