You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001663_01115
You are here: Home > Sequence: MGYG000001663_01115
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp900760675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp900760675 | |||||||||||
| CAZyme ID | MGYG000001663_01115 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 48216; End: 49691 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 94 | 252 | 7.7e-32 | 0.39211136890951276 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2723 | BglB | 1.14e-09 | 79 | 176 | 70 | 170 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| pfam01229 | Glyco_hydro_39 | 6.71e-07 | 97 | 204 | 70 | 190 | Glycosyl hydrolases family 39. |
| COG3693 | XynA | 8.86e-06 | 70 | 231 | 48 | 217 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| smart00633 | Glyco_10 | 8.89e-04 | 91 | 175 | 3 | 86 | Glycosyl hydrolase family 10. |
| pfam02449 | Glyco_hydro_42 | 0.001 | 79 | 111 | 21 | 54 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL15167.1 | 5.29e-242 | 22 | 491 | 26 | 496 |
| BBL03131.1 | 2.15e-241 | 22 | 491 | 26 | 496 |
| QNN22472.1 | 2.61e-172 | 24 | 479 | 29 | 477 |
| QNN22723.1 | 5.16e-110 | 34 | 474 | 32 | 475 |
| AEI51083.1 | 7.17e-107 | 38 | 473 | 43 | 484 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ZN2_A | 2.16e-13 | 79 | 368 | 44 | 339 | Glycosylhydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_B Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_C Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_D Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5BX9_A | 2.16e-13 | 79 | 368 | 44 | 339 | Structureof PslG from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5BXA_A Structure of PslG from Pseudomonas aeruginosa in complex with mannose [Pseudomonas aeruginosa PAO1] |
| 5JVK_A | 3.50e-09 | 81 | 244 | 129 | 290 | Structuralinsights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_B Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_C Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus] |
| 4IPL_A | 2.52e-06 | 70 | 176 | 77 | 189 | Thecrystal structure of 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae [Streptococcus pneumoniae TIGR4],4IPL_B The crystal structure of 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae [Streptococcus pneumoniae TIGR4],4IPN_B The complex structure of 6-phospho-beta-glucosidase BglA-2 with thiocellobiose-6P from Streptococcus pneumoniae [Streptococcus pneumoniae TIGR4],4IPN_E The complex structure of 6-phospho-beta-glucosidase BglA-2 with thiocellobiose-6P from Streptococcus pneumoniae [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P42973 | 5.91e-06 | 70 | 275 | 66 | 284 | Aryl-phospho-beta-D-glucosidase BglA OS=Bacillus subtilis (strain 168) OX=224308 GN=bglA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
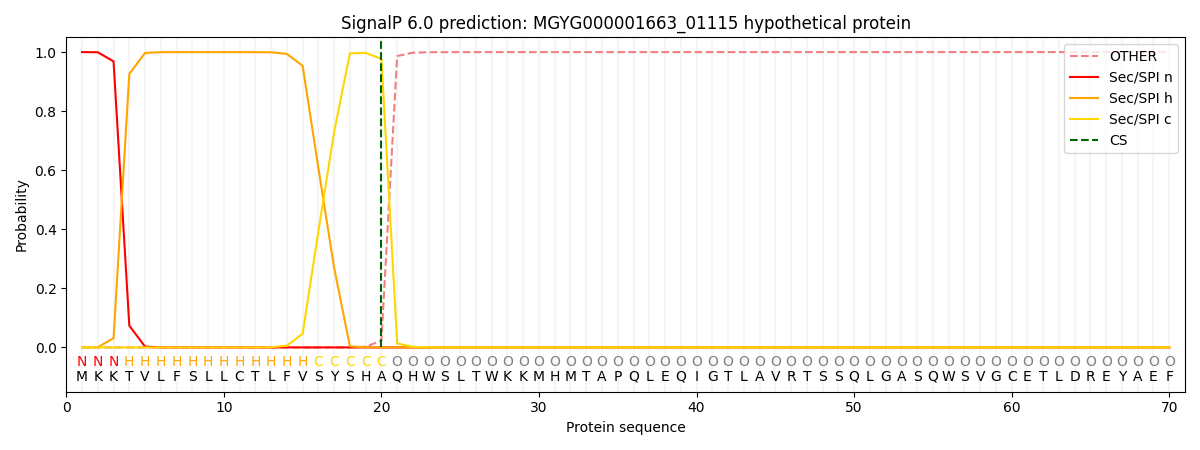
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000335 | 0.998993 | 0.000196 | 0.000161 | 0.000154 | 0.000146 |
