You are browsing environment: HUMAN GUT
MGYG000001665_01081
Basic Information
help
Species
UMGS1322 sp900550765
Lineage
Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; QALW01; UMGS1322; UMGS1322 sp900550765
CAZyme ID
MGYG000001665_01081
CAZy Family
GH140
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
886
97056.22
6.3772
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001665
2160920
MAG
United States
North America
Gene Location
Start: 2003;
End: 4663
Strand: -
No EC number prediction in MGYG000001665_01081.
Family
Start
End
Evalue
family coverage
GH140
398
859
4.5e-28
0.9611650485436893
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam13204
DUF4038
3.52e-31
404
766
1
320
Protein of unknown function (DUF4038). A family of putative cellulases.
more
pfam16586
DUF5060
1.29e-11
303
368
4
70
Domain of unknown function (DUF5060). This is the N-terminal domain of a putative glycoside hydrolase, DUF4038. It is found in a number of different bacterial orders.
more
PHA03270
PHA03270
8.39e-04
9
76
9
79
envelope glycoprotein C; Provisional
more
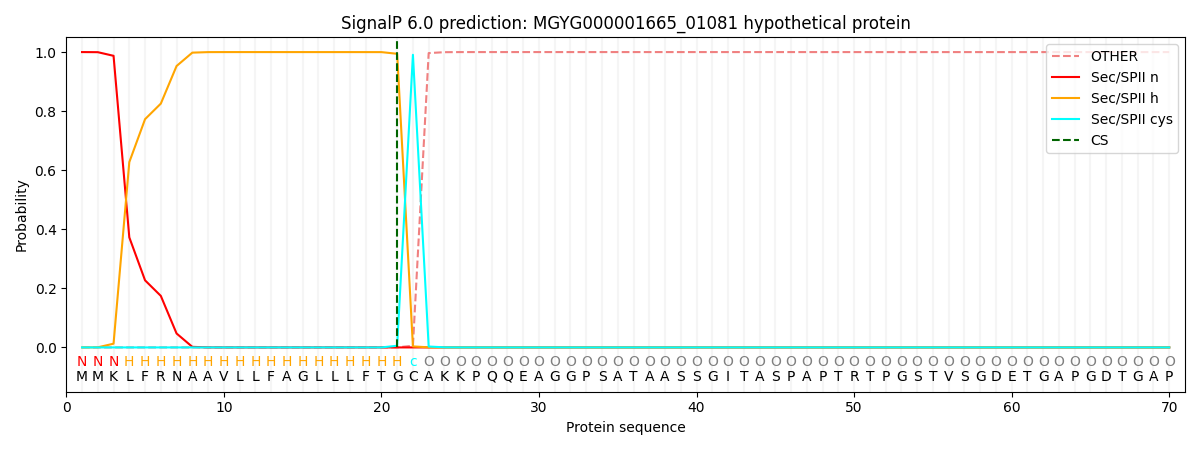
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000000
0.000001
1.000061
0.000000
0.000000
0.000000
There is no transmembrane helices in MGYG000001665_01081.