You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001666_01663
You are here: Home > Sequence: MGYG000001666_01663
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
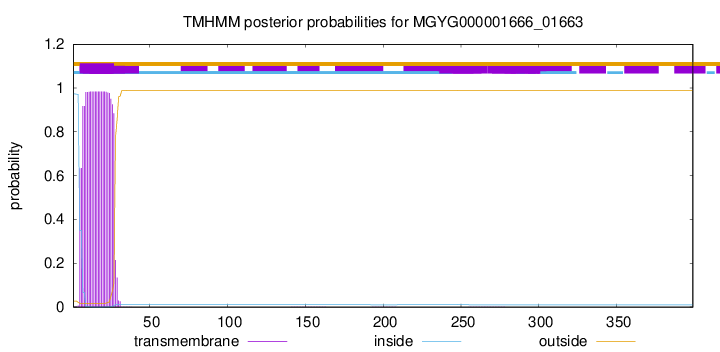
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; | |||||||||||
| CAZyme ID | MGYG000001666_01663 | |||||||||||
| CAZy Family | GH171 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2821; End: 4020 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH171 | 53 | 397 | 5.9e-111 | 0.9971830985915493 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07075 | DUF1343 | 1.15e-167 | 54 | 397 | 1 | 362 | Protein of unknown function (DUF1343). This family consists of several hypothetical bacterial proteins of around 400 residues in length. The function of this family is unknown. |
| COG3876 | YbbC | 1.73e-89 | 1 | 397 | 1 | 409 | Uncharacterized conserved protein YbbC, DUF1343 family [Function unknown]. |
| cd21002 | IgC1_MHC_II_beta_HLA-DM | 0.006 | 217 | 248 | 38 | 75 | Class II major histocompatibility complex (MHC) beta chain immunoglobulin domain of histocompatibility antigen (HLA) DM; member of the C1-set of Ig superfamily (IgSF) domains. The members here are composed of the Class II major histocompatibility complex (MHC) beta chain immunoglobulin domain of histocompatibility antigen (HLA) DM. Human HLA-DM plays a critical role in antigen presentation to CD4 T cells by catalyzing the exchange of peptides bound to MHC class II molecules. Type 1 diabetes is correlated with DM activation and it is also implicated in viral infections such as herpes simplex virus, celiac disease, multiple sclerosis, other autoimmune diseases, and leukemia. MHC class II molecules play a key role in the initiation of the antigen-specific immune reponse. These molecules have been shown to be expressed constitutively on the cell surface of professional antigen-presenting cells (APCs), including B-lymphocytes, monocytes, and macrophages in both humans and mice. The expression of these molecules has been shown to be induced in nonprofessional APCs such as keratinocyctes, and they are expressed on the surface of activated human T cells and on T cells from other species. The MHC II molecules present antigenic peptides to CD4(+) T-lymphocytes. These peptides derive mostly from proteolytic processing via the endocytic pathway, of antigens internalized by the APC. These peptides bind to the MHC class II molecules in the endosome before they are transported to the cell surface. MHC class II molecules are heterodimers, comprised of two similarly-sized membrane-spanning chains, alpha and beta. Each chain had two globular domains (N- and C-terminal), and a membrane-anchoring transmembrane segment. The two chains form a compact four-domain structure. The peptide-binding site is a cleft in the structure. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCQ31137.1 | 2.61e-185 | 22 | 399 | 12 | 388 |
| QCQ44443.1 | 2.61e-185 | 22 | 399 | 12 | 388 |
| QLK81838.1 | 2.61e-185 | 22 | 399 | 12 | 388 |
| QRM69467.1 | 2.61e-185 | 22 | 399 | 12 | 388 |
| QCQ35550.1 | 2.61e-185 | 22 | 399 | 12 | 388 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4JJA_A | 6.84e-173 | 33 | 399 | 5 | 370 | Crystalstructure of a DUF1343 family protein (BF0379) from Bacteroides fragilis NCTC 9343 at 1.30 A resolution [Bacteroides fragilis NCTC 9343] |
| 4K05_A | 9.82e-45 | 48 | 397 | 28 | 398 | Crystalstructure of a DUF1343 family protein (BF0371) from Bacteroides fragilis NCTC 9343 at 1.65 A resolution [Bacteroides fragilis NCTC 9343],4K05_B Crystal structure of a DUF1343 family protein (BF0371) from Bacteroides fragilis NCTC 9343 at 1.65 A resolution [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40407 | 7.66e-70 | 43 | 397 | 47 | 414 | Uncharacterized protein YbbC OS=Bacillus subtilis (strain 168) OX=224308 GN=ybbC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
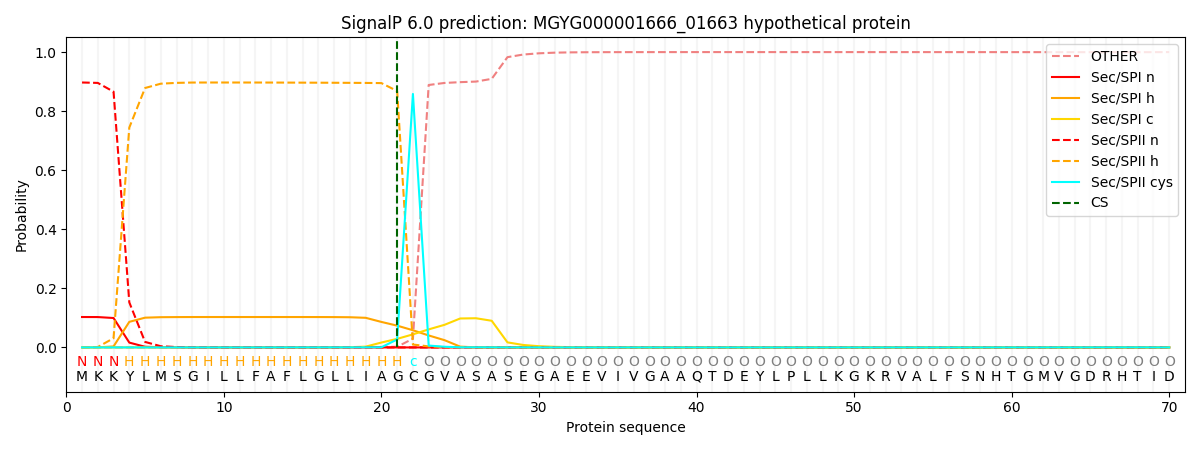
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000120 | 0.100812 | 0.898962 | 0.000049 | 0.000053 | 0.000040 |