You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001671_00597
You are here: Home > Sequence: MGYG000001671_00597
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | HGM11507 sp900761005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; UMGS1840; UMGS1840; HGM11507; HGM11507 sp900761005 | |||||||||||
| CAZyme ID | MGYG000001671_00597 | |||||||||||
| CAZy Family | PL15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7426; End: 10842 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL15 | 499 | 643 | 1.5e-32 | 0.9850746268656716 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16332 | DUF4962 | 9.28e-20 | 56 | 389 | 21 | 401 | Domain of unknown function (DUF4962). This family consists of uncharacterized proteins around 870 residues in length and is mainly found in various Bacteroides species. The function of this protein is unknown. |
| pfam07940 | Hepar_II_III | 1.80e-09 | 482 | 694 | 3 | 199 | Heparinase II/III-like protein. This family features sequences that are similar to a region of the Flavobacterium heparinum proteins heparinase II and heparinase III. The former is known to degrade heparin and heparin sulphate, whereas the latter predominantly degrades heparin sulphate. Both are secreted into the periplasmic space upon induction with heparin. |
| cd04086 | CBM35_mannanase-like | 1.08e-08 | 1004 | 1131 | 1 | 116 | Carbohydrate Binding Module 35 (CBM35); appended to several carbohydrate binding enzymes, including several glycoside hydrolase (GH) family 26 mannanase domains. This family includes carbohydrate binding module 35 (CBM35) domains that are appended to several carbohydrate binding enzymes, including periplasmic component of ABC-type sugar transport system involved in carbohydrate transport and metabolism, and several glycoside hydrolase (GH) domains, including GH26. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB35s belonging to this family are mannanase A from Clostridium thermocellum (GH26), Man26B from Paenibacillus sp. BME-14 (GH26), and the multifunctional Cel44C-Man26A from Paenibacillus polymyxa GS01 (which has two GH domains, GH44 and GH26). GH26 mainly includes mannan endo-1,4-beta-mannosidase which hydrolyzes 1,4-beta-D-linkages in mannans, galacto-mannans, glucomannans, and galactoglucomannans, but displays little activity towards other plant cell wall polysaccharides. A few proteins belonging to this family have additional CBM3 domains; these CBM3s are not found in the CBM6-CBM35-CBM36_like superfamily. |
| cd04083 | CBM35_Lmo2446-like | 3.51e-06 | 1007 | 1131 | 4 | 123 | Carbohydrate Binding Module 35 (CBM35) domains similar to Lmo2446. This family includes carbohydrate binding module 35 (CBM35) domains that are appended to several carbohydrate binding enzymes. Some CBM35 domains belonging to this family are appended to glycoside hydrolase (GH) family domains, including glycoside hydrolase family 31 (GH31), for example the CBM35 domain of Lmo2446, an uncharacterized protein from Listeria monocytogenes EGD-e. These CBM35s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. GH31 has a wide range of hydrolytic activities such as alpha-glucosidase, alpha-xylosidase, 6-alpha-glucosyltransferase, or alpha-1,4-glucan lyase, cleaving a terminal carbohydrate moiety from a substrate that may be a starch or a glycoprotein. Most characterized GH31 enzymes are alpha-glucosidases. |
| pfam16990 | CBM_35 | 1.49e-05 | 1022 | 1135 | 16 | 119 | Carbohydrate binding module (family 35). This is a mannan-specific carbohydrate binding domain, previously known as the X4 module. Unlike other carbohydrate binding modules, binding to substrate causes a conformational change. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNK58733.1 | 2.01e-196 | 6 | 1138 | 12 | 1134 |
| ASV74042.1 | 6.75e-61 | 43 | 674 | 45 | 681 |
| CDN57294.1 | 4.09e-60 | 55 | 681 | 33 | 679 |
| ABB73559.1 | 1.77e-59 | 53 | 702 | 56 | 700 |
| QUW23303.1 | 2.52e-58 | 46 | 775 | 20 | 768 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3A0O_A | 1.88e-53 | 46 | 655 | 26 | 657 | Crystalstructure of alginate lyase from Agrobacterium tumefaciens C58 [Agrobacterium fabrum str. C58],3A0O_B Crystal structure of alginate lyase from Agrobacterium tumefaciens C58 [Agrobacterium fabrum str. C58] |
| 3AFL_A | 3.50e-52 | 46 | 655 | 26 | 657 | Crystalstructure of exotype alginate lyase Atu3025 H531A complexed with alginate trisaccharide [Agrobacterium fabrum str. C58] |
| 6LJA_A | 3.00e-23 | 57 | 655 | 52 | 697 | ChainA, Heparinase II/III-like protein [Bacteroides intestinalis DSM 17393] |
| 6LJL_A | 5.48e-21 | 57 | 655 | 52 | 697 | ChainA, Heparinase II/III-like protein [Bacteroides intestinalis DSM 17393] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
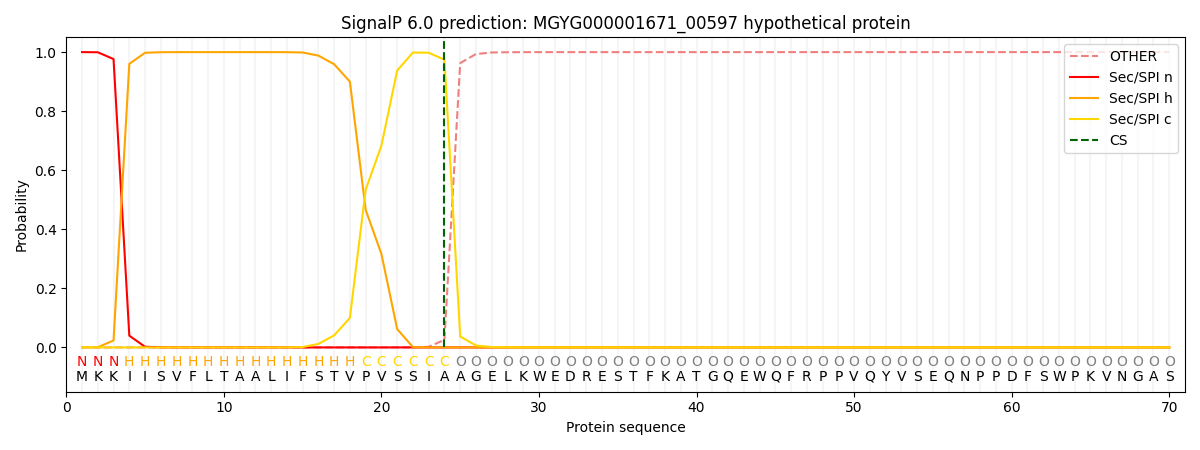
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000236 | 0.999046 | 0.000182 | 0.000177 | 0.000165 | 0.000149 |
