You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001672_00683
You are here: Home > Sequence: MGYG000001672_00683
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS995 sp900547465 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; UMGS995; UMGS995 sp900547465 | |||||||||||
| CAZyme ID | MGYG000001672_00683 | |||||||||||
| CAZy Family | CE3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 41029; End: 43707 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH25 | 701 | 880 | 1.6e-27 | 0.9887005649717514 |
| CE3 | 219 | 363 | 1.2e-16 | 0.6958762886597938 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06414 | GH25_LytC-like | 3.42e-40 | 701 | 891 | 5 | 191 | The LytC lysozyme of Streptococcus pneumoniae is a bacterial cell wall hydrolase that cleaves the beta1-4-glycosydic bond located between the N-acetylmuramoyl-N-glucosaminyl residues of the cell wall polysaccharide chains. LytC is composed of a C-terminal glycosyl hydrolase family 25 (GH25) domain and an N-terminal choline-binding module (CBM) consisting of eleven homologous repeats that specifically recognizes the choline residues of pneumococcal lipoteichoic and teichoic acids. This domain arrangement is the reverse of the major pneumococcal autolysin, LytA, and the CPL-1-like lytic enzymes of the pneumococcal bacteriophages, in which the CBM (consisting of six repeats) is at the C-terminus. This model represents the C-terminal catalytic domain of the LytC-like enzymes. |
| cd01841 | NnaC_like | 3.15e-36 | 193 | 369 | 1 | 174 | NnaC (CMP-NeuNAc synthetase) _like subfamily of SGNH_hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles two of the three components of typical Ser-His-Asp(Glu) triad from other serine hydrolases. E. coli NnaC appears to be involved in polysaccharide synthesis. |
| cd01828 | sialate_O-acetylesterase_like2 | 1.12e-32 | 197 | 370 | 4 | 169 | sialate_O-acetylesterase_like subfamily of the SGNH-hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| pfam13472 | Lipase_GDSL_2 | 1.58e-25 | 197 | 360 | 1 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| cd06523 | GH25_PlyB-like | 1.27e-18 | 699 | 888 | 2 | 175 | PlyB is a bacteriophage endolysin that displays potent lytic activity toward Bacillus anthracis. PlyB has an N-terminal glycosyl hydrolase family 25 (GH25) catalytic domain and a C-terminal bacterial SH3-like domain, SH3b. Both domains are required for effective catalytic activity. Endolysins are produced by bacteriophages at the end of their life cycle and participate in lysing the bacterial cell in order to release the newly formed progeny. Endolysins (also referred to as endo-N-acetylmuramidases or peptidoglycan hydrolases) degrade bacterial cell walls by catalyzing the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJA04626.1 | 2.63e-28 | 695 | 890 | 108 | 313 |
| QQR24886.1 | 4.67e-28 | 695 | 890 | 108 | 313 |
| ANU71242.1 | 4.67e-28 | 695 | 890 | 108 | 313 |
| QIX10955.1 | 4.67e-28 | 695 | 890 | 108 | 313 |
| ASU20273.1 | 4.67e-28 | 695 | 890 | 108 | 313 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2WW5_A | 8.16e-28 | 680 | 891 | 251 | 468 | 3D-structureof the modular autolysin LytC from Streptococcus pneumoniae at 1.6 A resolution [Streptococcus pneumoniae R6],2WWD_A 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with pneummococcal peptidoglycan fragment [Streptococcus pneumoniae R6] |
| 2WWC_A | 1.96e-27 | 680 | 891 | 251 | 468 | 3D-structureof the modular autolysin LytC from Streptococcus pneumoniae in complex with synthetic peptidoglycan ligand [Streptococcus pneumoniae R6] |
| 2HSJ_A | 2.13e-19 | 178 | 369 | 20 | 213 | Thestructure of a putative platelet activating factor from Streptococcus pneumonia. [Streptococcus pneumoniae TIGR4],2HSJ_B The structure of a putative platelet activating factor from Streptococcus pneumonia. [Streptococcus pneumoniae TIGR4],2HSJ_C The structure of a putative platelet activating factor from Streptococcus pneumonia. [Streptococcus pneumoniae TIGR4],2HSJ_D The structure of a putative platelet activating factor from Streptococcus pneumonia. [Streptococcus pneumoniae TIGR4] |
| 6XPM_A | 6.28e-16 | 197 | 370 | 30 | 202 | ChainA, Lysophospholipase L1 [Phocaeicola vulgatus] |
| 6NJC_A | 2.89e-15 | 197 | 370 | 30 | 202 | ChainA, Sialate O-acetylesterase [Phocaeicola vulgatus ATCC 8482],6NJC_B Chain B, Sialate O-acetylesterase [Phocaeicola vulgatus ATCC 8482],6XPG_A Chain A, Lysophospholipase L1 [Phocaeicola vulgatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P13266 | 8.19e-08 | 196 | 366 | 253 | 419 | N-acylneuraminate cytidylyltransferase OS=Escherichia coli OX=562 GN=neuA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
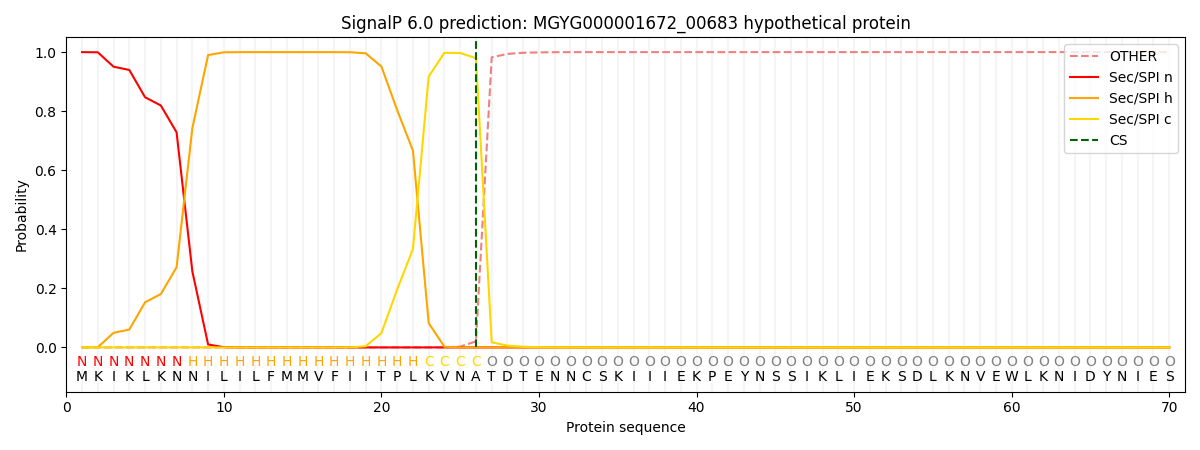
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000260 | 0.999092 | 0.000183 | 0.000144 | 0.000137 | 0.000130 |
