You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001673_00547
You are here: Home > Sequence: MGYG000001673_00547
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
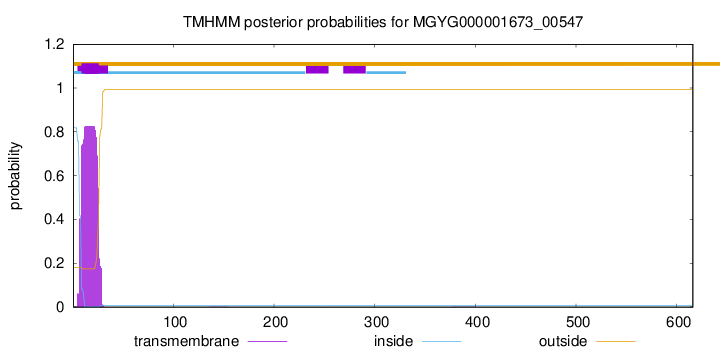
TMHMM annotations
Basic Information help
| Species | UMGS1537 sp900552695 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; UBA1212; UBA1255; UMGS1537; UMGS1537 sp900552695 | |||||||||||
| CAZyme ID | MGYG000001673_00547 | |||||||||||
| CAZy Family | GH163 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13282; End: 15132 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH163 | 245 | 507 | 1.6e-93 | 0.9960159362549801 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16126 | DUF4838 | 2.51e-112 | 238 | 507 | 1 | 263 | Domain of unknown function (DUF4838). This family consists of several uncharacterized proteins found in various Bacteroides and Chloroflexus species. The function of this family is unknown. |
| pfam03648 | Glyco_hydro_67N | 7.13e-08 | 59 | 152 | 24 | 119 | Glycosyl hydrolase family 67 N-terminus. Alpha-glucuronidases, components of an ensemble of enzymes central to the recycling of photosynthetic biomass, remove the alpha-1,2 linked 4-O-methyl glucuronic acid from xylans. This family represents the N-terminal region of alpha-glucuronidase. The N-terminal domain forms a two-layer sandwich, each layer being formed by a beta sheet of five strands. A further two helices form part of the interface with the central, catalytic, module (pfam07488). |
| cd11288 | gelsolin_S5_like | 0.008 | 60 | 108 | 46 | 92 | Gelsolin sub-domain 5-like domain found in gelsolin, severin, villin, and related proteins. Gelsolin repeats occur in gelsolin, severin, villin, advillin, villidin, supervillin, flightless, quail, fragmin, and other proteins, usually in several copies. They co-occur with villin headpiece domains, leucine-rich repeats, and several other domains. These gelsolin-related actin binding proteins (GRABPs) play regulatory roles in the assembly and disassembly of actin filaments; they are involved in F-actin capping, uncapping, severing, or the nucleation of actin filaments. Severing of actin filaments is Ca2+ dependent. Villins are also linked to generating bundles of F-actin with uniform filament polarity, which is most likely mediated by their extra villin headpiece domain. Many family members have also adopted functions in the nucleus, including the regulation of transcription. Supervillin, gelsolin, and flightless I are involved in intracellular signaling via nuclear hormone receptors. The gelsolin-like domain is distantly related to the actin depolymerizing domains found in cofilin and similar proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOY90686.1 | 4.15e-116 | 39 | 572 | 10 | 528 |
| ASV73444.1 | 6.92e-107 | 52 | 554 | 32 | 522 |
| AIF26900.1 | 8.96e-107 | 52 | 500 | 10 | 468 |
| AIE83908.1 | 4.61e-105 | 57 | 508 | 22 | 452 |
| QQL44163.1 | 9.49e-104 | 54 | 554 | 19 | 508 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
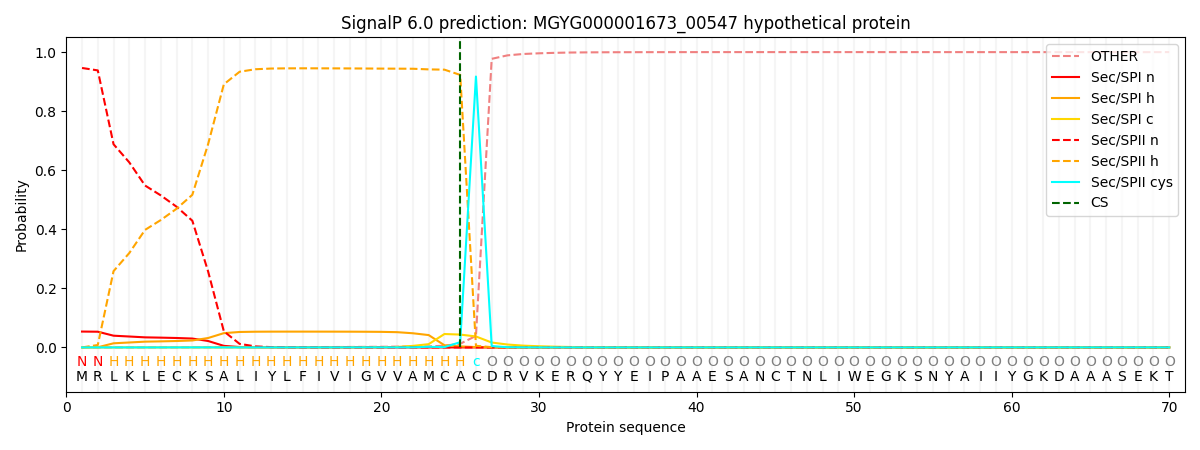
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000193 | 0.052039 | 0.947705 | 0.000021 | 0.000034 | 0.000024 |