You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001673_01911
You are here: Home > Sequence: MGYG000001673_01911
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
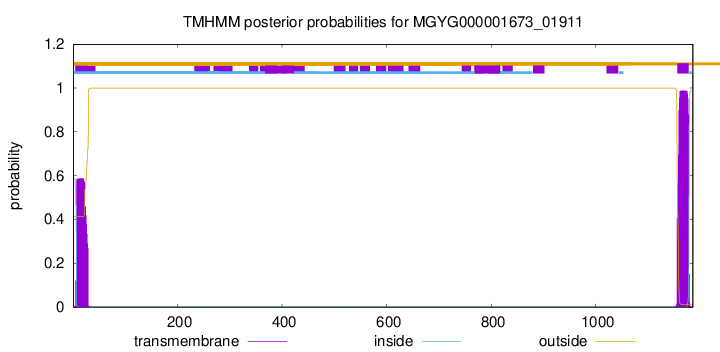
TMHMM annotations
Basic Information help
| Species | UMGS1537 sp900552695 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; UBA1212; UBA1255; UMGS1537; UMGS1537 sp900552695 | |||||||||||
| CAZyme ID | MGYG000001673_01911 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9877; End: 13437 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 684 | 924 | 4.7e-23 | 0.5661538461538461 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 2.65e-21 | 628 | 924 | 77 | 367 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam12708 | Pectate_lyase_3 | 6.85e-04 | 629 | 690 | 4 | 58 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23464.1 | 3.20e-183 | 371 | 1055 | 71 | 745 |
| QCD41820.1 | 3.17e-115 | 418 | 992 | 248 | 834 |
| ADB48561.1 | 1.11e-41 | 624 | 1091 | 25 | 474 |
| QYH35737.1 | 4.20e-39 | 623 | 1124 | 122 | 631 |
| QGQ94767.1 | 3.38e-38 | 624 | 1103 | 34 | 507 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MXN_A | 8.42e-10 | 632 | 847 | 25 | 213 | Crystalstructure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_B Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_C Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_D Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184] |
| 5OLP_A | 5.85e-09 | 620 | 880 | 38 | 294 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
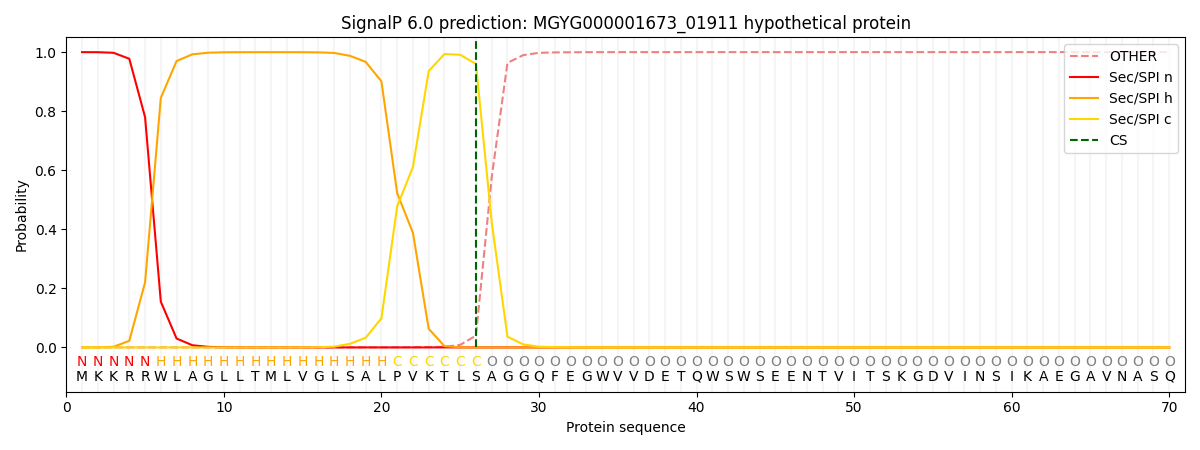
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000595 | 0.998256 | 0.000488 | 0.000258 | 0.000193 | 0.000173 |