You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001681_00557
You are here: Home > Sequence: MGYG000001681_00557
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus sp900761275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; Ruminococcus sp900761275 | |||||||||||
| CAZyme ID | MGYG000001681_00557 | |||||||||||
| CAZy Family | CBM23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7494; End: 11432 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 213 | 459 | 7.1e-78 | 0.9802371541501976 |
| GH5 | 916 | 1169 | 6.2e-71 | 0.9762845849802372 |
| CBM23 | 32 | 175 | 8.1e-30 | 0.9074074074074074 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 7.26e-30 | 916 | 1168 | 3 | 272 | Cellulase (glycosyl hydrolase family 5). |
| pfam00150 | Cellulase | 1.90e-25 | 212 | 458 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| pfam03425 | CBM_11 | 2.27e-07 | 33 | 144 | 7 | 124 | Carbohydrate binding domain (family 11). |
| cd14256 | Dockerin_I | 2.39e-04 | 1236 | 1300 | 2 | 55 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKS47268.1 | 5.11e-118 | 205 | 867 | 414 | 1235 |
| CCO04360.1 | 5.65e-89 | 205 | 501 | 41 | 335 |
| QJU43361.1 | 6.22e-88 | 902 | 1265 | 37 | 402 |
| QGH23260.1 | 6.22e-88 | 902 | 1265 | 37 | 402 |
| QCJ05579.1 | 6.22e-88 | 902 | 1265 | 37 | 402 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1WKY_A | 1.72e-84 | 205 | 676 | 11 | 459 | Crystalstructure of alkaline mannanase from Bacillus sp. strain JAMB-602: catalytic domain and its Carbohydrate Binding Module [Bacillus sp. JAMB-602] |
| 3JUG_A | 1.30e-82 | 899 | 1203 | 18 | 312 | Crystalstructure of endo-beta-1,4-mannanase from the alkaliphilic Bacillus sp. N16-5 [Bacillus sp. N16-5] |
| 2WHJ_A | 2.20e-80 | 905 | 1203 | 2 | 290 | Understandinghow diverse mannanases recognise heterogeneous substrates [Salipaludibacillus agaradhaerens] |
| 2WHL_A | 4.95e-80 | 905 | 1203 | 1 | 287 | Understandinghow diverse mannanases recognise heterogeneous substrates [Salipaludibacillus agaradhaerens] |
| 1BQC_A | 1.80e-60 | 204 | 496 | 4 | 298 | Beta-MannanaseFrom Thermomonospora Fusca [Thermobifida fusca],2MAN_A Mannotriose Complex Of Thermomonospora Fusca Beta-Mannanase [Thermobifida fusca],3MAN_A Mannohexaose Complex Of Thermomonospora Fusca Beta-mannanase [Thermobifida fusca] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G1K3N4 | 1.21e-79 | 905 | 1203 | 2 | 290 | Mannan endo-1,4-beta-mannosidase OS=Salipaludibacillus agaradhaerens OX=76935 PE=1 SV=1 |
| B3PF24 | 6.07e-56 | 198 | 501 | 43 | 347 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=man5A PE=1 SV=1 |
| P51529 | 1.31e-54 | 898 | 1203 | 32 | 329 | Mannan endo-1,4-beta-mannosidase OS=Streptomyces lividans OX=1916 GN=manA PE=1 SV=2 |
| P22533 | 1.73e-50 | 223 | 492 | 55 | 326 | Beta-mannanase/endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=manA PE=1 SV=2 |
| Q53317 | 5.25e-19 | 1236 | 1309 | 439 | 512 | Xylanase/beta-glucanase OS=Ruminococcus flavefaciens OX=1265 GN=xynD PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
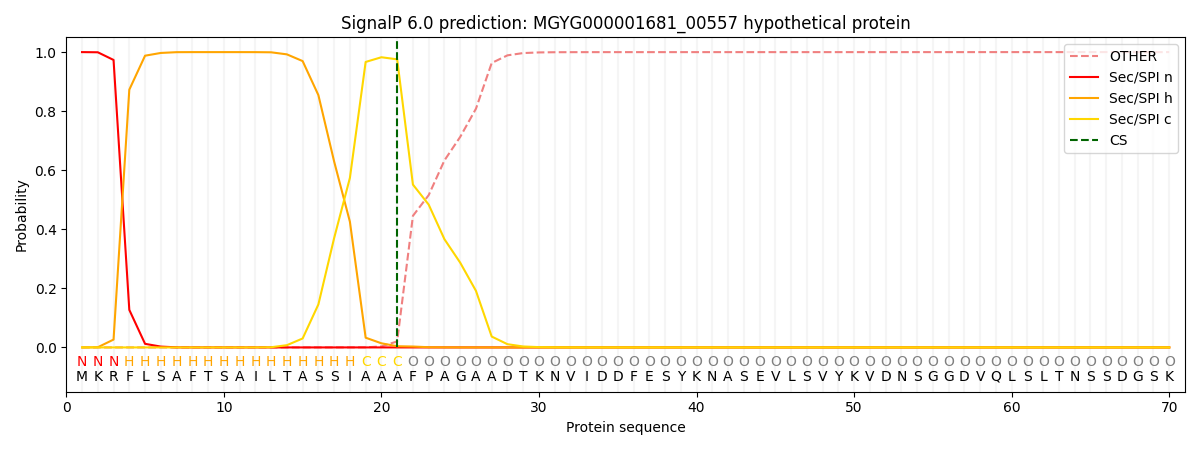
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000376 | 0.998867 | 0.000183 | 0.000197 | 0.000175 | 0.000160 |
