You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001681_01535
You are here: Home > Sequence: MGYG000001681_01535
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
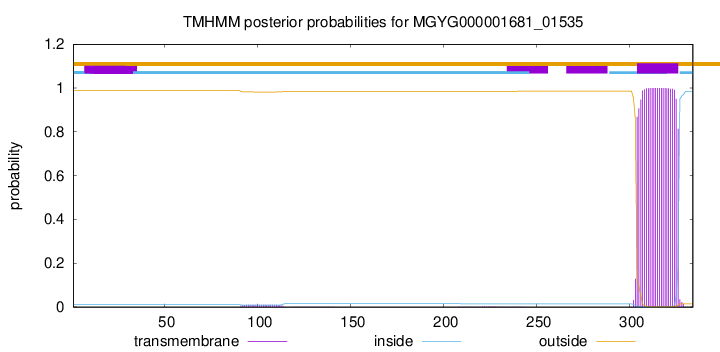
TMHMM annotations
Basic Information help
| Species | Ruminococcus sp900761275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; Ruminococcus sp900761275 | |||||||||||
| CAZyme ID | MGYG000001681_01535 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 258; End: 1262 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 22 | 225 | 8.1e-50 | 0.41866028708133973 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 1.47e-32 | 68 | 225 | 232 | 373 | Glycosyl hydrolase family 9. |
| PLN02345 | PLN02345 | 9.37e-05 | 128 | 155 | 361 | 388 | endoglucanase |
| PLN02909 | PLN02909 | 1.20e-04 | 127 | 180 | 381 | 453 | Endoglucanase |
| PLN00119 | PLN00119 | 3.64e-04 | 127 | 155 | 385 | 413 | endoglucanase |
| PLN02420 | PLN02420 | 0.001 | 127 | 155 | 403 | 431 | endoglucanase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADU21576.1 | 1.52e-119 | 4 | 332 | 576 | 900 |
| QNU67291.1 | 5.90e-82 | 4 | 239 | 562 | 801 |
| BAI52927.1 | 1.49e-79 | 1 | 228 | 570 | 800 |
| CBL17684.1 | 2.43e-79 | 9 | 232 | 627 | 861 |
| CUH93326.1 | 8.87e-79 | 1 | 228 | 562 | 789 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1UT9_A | 4.31e-65 | 1 | 231 | 377 | 606 | ChainA, CELLULOSE 1,4-BETA-CELLOBIOSIDASE [Acetivibrio thermocellus] |
| 1RQ5_A | 6.42e-64 | 1 | 228 | 377 | 603 | ChainA, Cellobiohydrolase [Acetivibrio thermocellus] |
| 6DHT_A | 1.04e-21 | 28 | 225 | 375 | 560 | Bacteroidesovatus GH9 Bacova_02649 [Bacteroides ovatus ATCC 8483] |
| 4CJ0_A | 1.07e-18 | 26 | 228 | 374 | 547 | ChainA, ENDOGLUCANASE D [Acetivibrio thermocellus],4CJ1_A Chain A, ENDOGLUCANASE D [Acetivibrio thermocellus] |
| 1CLC_A | 1.08e-18 | 26 | 228 | 388 | 561 | ChainA, ENDOGLUCANASE CELD; EC: 3.2.1.4 [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A3DCH1 | 2.12e-62 | 1 | 228 | 584 | 810 | Cellulose 1,4-beta-cellobiosidase OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celK PE=3 SV=1 |
| P0C2S1 | 5.57e-62 | 1 | 228 | 584 | 810 | Cellulose 1,4-beta-cellobiosidase OS=Acetivibrio thermocellus OX=1515 GN=celK PE=1 SV=1 |
| Q05156 | 6.11e-46 | 26 | 232 | 551 | 745 | Cellulase 1 OS=Streptomyces reticuli OX=1926 GN=cel1 PE=1 SV=1 |
| P14090 | 3.44e-39 | 26 | 232 | 713 | 910 | Endoglucanase C OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cenC PE=1 SV=2 |
| P10476 | 1.50e-38 | 26 | 228 | 405 | 597 | Endoglucanase A OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=celA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
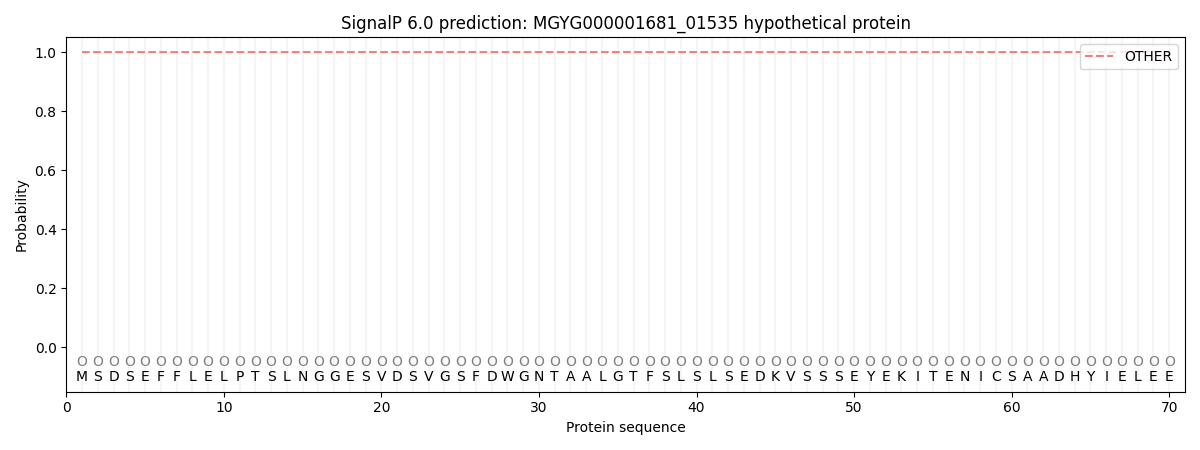
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000046 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |