You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001689_01583
You are here: Home > Sequence: MGYG000001689_01583
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
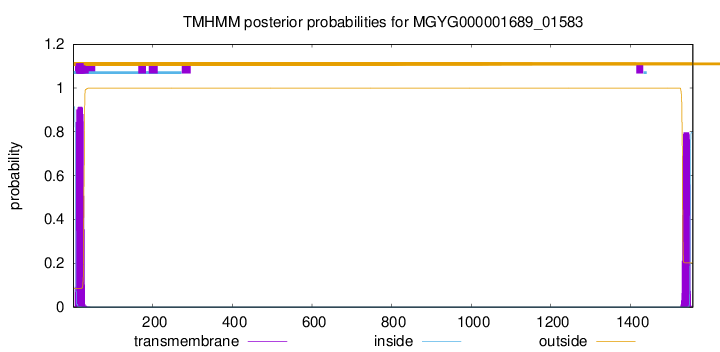
TMHMM annotations
Basic Information help
| Species | Blautia coccoides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia; Blautia coccoides | |||||||||||
| CAZyme ID | MGYG000001689_01583 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 45337; End: 50010 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 417 | 952 | 3.1e-134 | 0.9755600814663951 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PTZ00121 | PTZ00121 | 1.05e-06 | 1236 | 1525 | 1325 | 1590 | MAEBL; Provisional |
| pfam02018 | CBM_4_9 | 1.54e-06 | 34 | 166 | 1 | 130 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| pfam13229 | Beta_helix | 2.20e-06 | 557 | 770 | 1 | 154 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam02018 | CBM_4_9 | 5.81e-06 | 1070 | 1150 | 5 | 86 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| pfam08480 | Disaggr_assoc | 2.13e-05 | 743 | 917 | 9 | 194 | Disaggregatase related. This domain is found in disaggregatases and several hypothetical proteins of the archaeal genus Methanosarcina. Disaggregatases cause aggregates to separate into single cells and contain parallel beta-helix repeats. Also see pfam06848. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIB56278.1 | 0.0 | 1 | 1557 | 1 | 1557 |
| QMW80950.1 | 0.0 | 1 | 1557 | 8 | 1564 |
| QQQ95039.1 | 0.0 | 1 | 1557 | 1 | 1625 |
| ASU30251.1 | 0.0 | 1 | 1557 | 15 | 1639 |
| ANU77449.1 | 0.0 | 1 | 1557 | 15 | 1639 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 2.98e-39 | 415 | 922 | 6 | 551 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 6KQT_A | 4.07e-35 | 352 | 781 | 176 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 7V6I_A | 6.38e-34 | 419 | 917 | 15 | 578 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 6KQS_A | 2.21e-33 | 352 | 781 | 176 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
| 3OEB_A | 8.09e-06 | 1071 | 1212 | 7 | 143 | Crystalstructure of the Q121E mutant of C.polysaccharolyticus CBM16-1 bound to mannopentaose [Caldanaerobius polysaccharolyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 7.77e-11 | 1221 | 1423 | 1666 | 1872 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
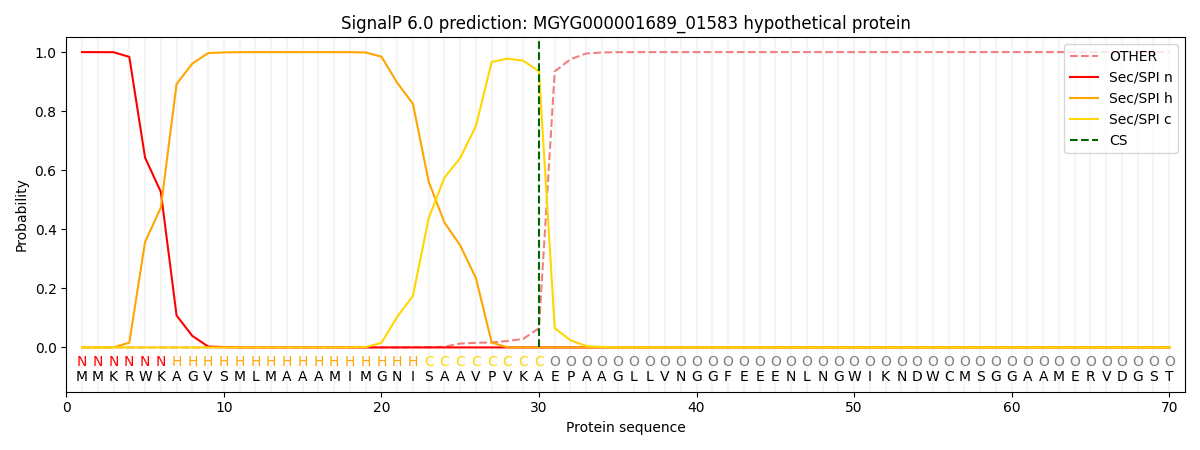
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000252 | 0.999078 | 0.000179 | 0.000184 | 0.000158 | 0.000139 |