You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001697_01418
You are here: Home > Sequence: MGYG000001697_01418
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
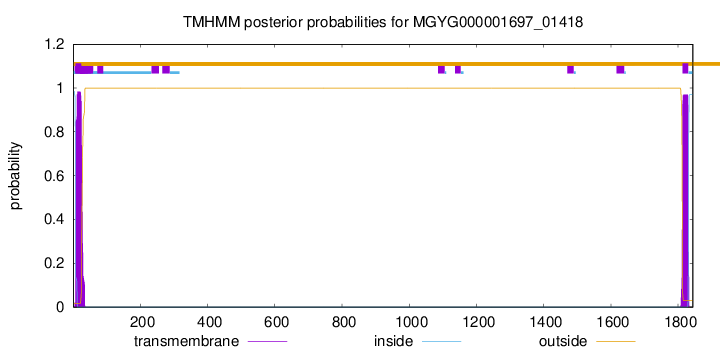
TMHMM annotations
Basic Information help
| Species | Erysipelatoclostridium saccharogumia | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Erysipelatoclostridium; Erysipelatoclostridium saccharogumia | |||||||||||
| CAZyme ID | MGYG000001697_01418 | |||||||||||
| CAZy Family | GH20 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4007; End: 9541 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH20 | 177 | 500 | 1.9e-96 | 0.9703264094955489 |
| GH29 | 752 | 1053 | 1.6e-76 | 0.8526011560693642 |
| CBM32 | 1475 | 1582 | 1.1e-25 | 0.8629032258064516 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06568 | GH20_SpHex_like | 4.40e-157 | 181 | 513 | 1 | 329 | A subgroup of the Glycosyl hydrolase family 20 (GH20) catalytic domain found in proteins similar to the N-acetylhexosaminidase from Streptomyces plicatus (SpHex). SpHex catalyzes the hydrolysis of N-acetyl-beta-hexosaminides. An Asp residue within the active site plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. Proteins belonging to this subgroup lack the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. |
| cd06563 | GH20_chitobiase-like | 1.87e-117 | 181 | 513 | 1 | 357 | The chitobiase of Serratia marcescens is a beta-N-1,4-acetylhexosaminidase with a glycosyl hydrolase family 20 (GH20) domain that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This GH20 domain family includes an N-acetylglucosamidase (GlcNAcase A) from Pseudoalteromonas piscicida and an N-acetylhexosaminidase (SpHex) from Streptomyces plicatus. SpHex lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| pfam00728 | Glyco_hydro_20 | 3.00e-97 | 181 | 500 | 1 | 345 | Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. |
| COG3669 | AfuC | 3.96e-84 | 743 | 1193 | 4 | 430 | Alpha-L-fucosidase [Carbohydrate transport and metabolism]. |
| COG3525 | Chb | 5.00e-80 | 36 | 537 | 131 | 649 | N-acetyl-beta-hexosaminidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYE34854.1 | 3.06e-229 | 742 | 1540 | 38 | 816 |
| QAS60248.1 | 3.06e-229 | 742 | 1540 | 38 | 816 |
| SLK21779.1 | 4.55e-228 | 723 | 1540 | 21 | 816 |
| ATD55836.1 | 4.55e-228 | 723 | 1540 | 21 | 816 |
| QBJ76136.1 | 4.55e-228 | 723 | 1540 | 21 | 816 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GH4_A | 3.01e-145 | 31 | 533 | 31 | 525 | Crystalstructure of beta-hexosaminidase from Paenibacillus sp. TS12 [Paenibacillus sp.],3GH5_A Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GlcNAc [Paenibacillus sp.],3GH7_A Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GalNAc [Paenibacillus sp.],3SUR_A Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with NAG-thiazoline. [Paenibacillus sp. TS12],3SUS_A Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with Gal-NAG-thiazoline [Paenibacillus sp. TS12],3SUT_A Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with PUGNAc [Paenibacillus sp. TS12],3SUU_A Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with Gal-PUGNAc [Paenibacillus sp. TS12],3SUV_A Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with NHAc-DNJ [Paenibacillus sp. TS12],3SUW_A Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with NHAc-CAS [Paenibacillus sp. TS12] |
| 6ORG_A | 3.31e-127 | 744 | 1194 | 5 | 449 | Crystalstructure of SpGH29 [Streptococcus pneumoniae TIGR4],6ORG_B Crystal structure of SpGH29 [Streptococcus pneumoniae TIGR4] |
| 6OR4_A | 4.07e-126 | 744 | 1194 | 5 | 449 | Crystalstructure of SpGH29 [Streptococcus pneumoniae TIGR4],6OR4_B Crystal structure of SpGH29 [Streptococcus pneumoniae TIGR4],6ORH_A Crystal structure of SpGH29 [Streptococcus pneumoniae TIGR4],6ORH_B Crystal structure of SpGH29 [Streptococcus pneumoniae TIGR4] |
| 6ORF_A | 4.21e-126 | 744 | 1194 | 5 | 449 | Crystalstructure of SpGH29 [Streptococcus pneumoniae TIGR4],6ORF_B Crystal structure of SpGH29 [Streptococcus pneumoniae TIGR4] |
| 1HP4_A | 7.32e-115 | 41 | 533 | 27 | 509 | ChainA, Beta-n-acetylhexosaminidase [Streptomyces plicatus],1HP5_A Chain A, Beta-n-acetylhexosaminidase [Streptomyces plicatus],1JAK_A Chain A, Beta-N-acetylhexosaminidase [Streptomyces plicatus],1M01_A Chain A, Beta-N-acetylhexosaminidase [Streptomyces plicatus],5FCZ_A Chain A, B-N-acetylhexosaminidase [Streptomyces plicatus],5FD0_A Chain A, B-N-acetylhexosaminidase [Streptomyces plicatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q7XUR3 | 1.32e-120 | 748 | 1193 | 38 | 476 | Putative alpha-L-fucosidase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=Os04g0560400 PE=3 SV=2 |
| Q8GW72 | 1.18e-116 | 748 | 1193 | 36 | 477 | Alpha-L-fucosidase 1 OS=Arabidopsis thaliana OX=3702 GN=FUC1 PE=1 SV=2 |
| Q7WUL4 | 1.40e-78 | 40 | 537 | 5 | 488 | Beta-N-acetylhexosaminidase OS=Cellulomonas fimi OX=1708 GN=hex20 PE=1 SV=1 |
| P49008 | 1.42e-74 | 41 | 528 | 37 | 533 | Beta-hexosaminidase OS=Porphyromonas gingivalis (strain ATCC BAA-308 / W83) OX=242619 GN=nahA PE=3 SV=2 |
| B2UQG6 | 3.55e-57 | 98 | 527 | 75 | 529 | Beta-hexosaminidase Amuc_0868 OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=Amuc_0868 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
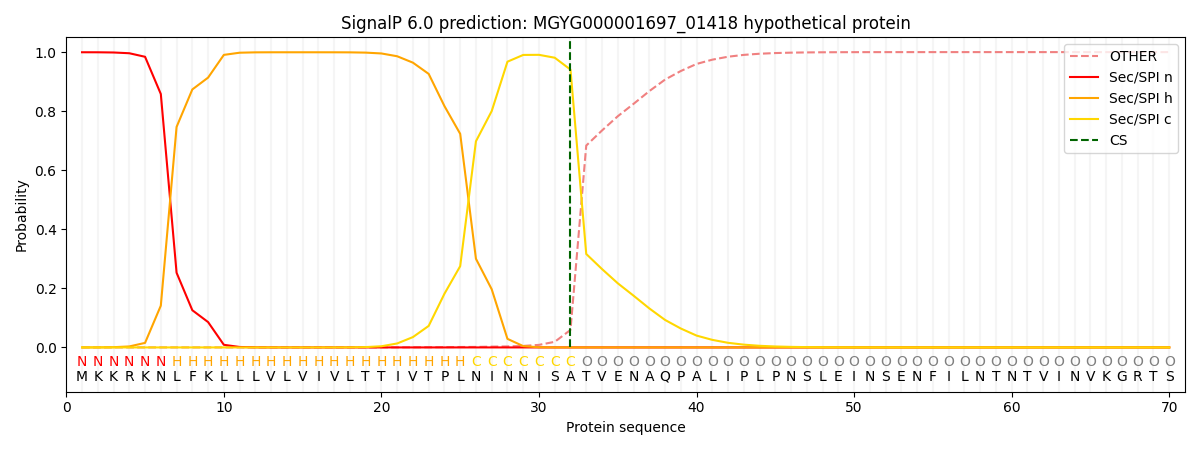
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000677 | 0.997812 | 0.000933 | 0.000203 | 0.000190 | 0.000168 |