You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001697_02156
You are here: Home > Sequence: MGYG000001697_02156
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
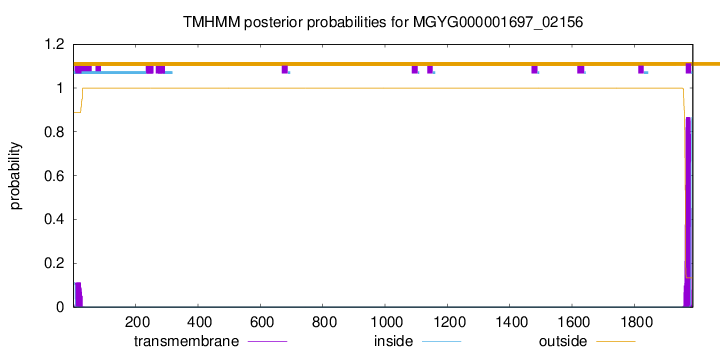
TMHMM annotations
Basic Information help
| Species | Erysipelatoclostridium saccharogumia | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Erysipelatoclostridium; Erysipelatoclostridium saccharogumia | |||||||||||
| CAZyme ID | MGYG000001697_02156 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14864; End: 20830 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 1404 | 1531 | 2.7e-19 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.31e-17 | 1401 | 1532 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 1.58e-07 | 297 | 427 | 3 | 116 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.57e-06 | 1397 | 1523 | 9 | 134 | Substituted updates: Jan 31, 2002 |
| pfam00754 | F5_F8_type_C | 1.62e-06 | 138 | 254 | 17 | 116 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 5.63e-06 | 325 | 425 | 41 | 130 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXH52486.1 | 0.0 | 282 | 1642 | 108 | 1463 |
| ALG48827.1 | 0.0 | 282 | 1642 | 108 | 1463 |
| ATD48595.1 | 0.0 | 282 | 1642 | 108 | 1463 |
| AOY54034.1 | 0.0 | 282 | 1642 | 108 | 1463 |
| SQG39183.1 | 0.0 | 282 | 1642 | 108 | 1463 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4A41_A | 3.22e-12 | 1399 | 1534 | 29 | 159 | CpGH89CBM32-5,from Clostridium perfringens, in complex with galactose [Clostridium perfringens],4A44_A CpGH89CBM32-5, from Clostridium perfringens, in complex with the Tn Antigen [Clostridium perfringens],4A45_A CpGH89CBM32-5, from Clostridium perfringens, in complex with GalNAc- beta-1,3-galactose [Clostridium perfringens],4AAX_A CpGH89CBM32-5, from Clostridium perfringens, in complex with N- acetylgalactosamine [Clostridium perfringens] |
| 2V72_A | 1.15e-10 | 1401 | 1534 | 17 | 141 | Thestructure of the family 32 CBM from C. perfringens NanJ in complex with galactose [Clostridium perfringens] |
| 1EUT_A | 2.16e-07 | 1406 | 1534 | 474 | 601 | Sialidase,Large 68kd Form, Complexed With Galactose [Micromonospora viridifaciens],1EUU_A Sialidase Or Neuraminidase, Large 68kd Form [Micromonospora viridifaciens] |
| 1W8N_A | 2.16e-07 | 1406 | 1534 | 470 | 597 | Contributionof the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. [Micromonospora viridifaciens],1W8O_A Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens [Micromonospora viridifaciens] |
| 1WCQ_A | 2.16e-07 | 1406 | 1534 | 470 | 597 | Mutagenesisof the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_B Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_C Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 3.65e-13 | 1749 | 1978 | 1667 | 1914 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
| P0DTR4 | 1.53e-07 | 1336 | 1534 | 442 | 643 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| Q02834 | 1.23e-06 | 1406 | 1534 | 516 | 643 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P29767 | 1.48e-06 | 1420 | 1569 | 72 | 217 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
| P26831 | 2.15e-06 | 1711 | 1871 | 1389 | 1561 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
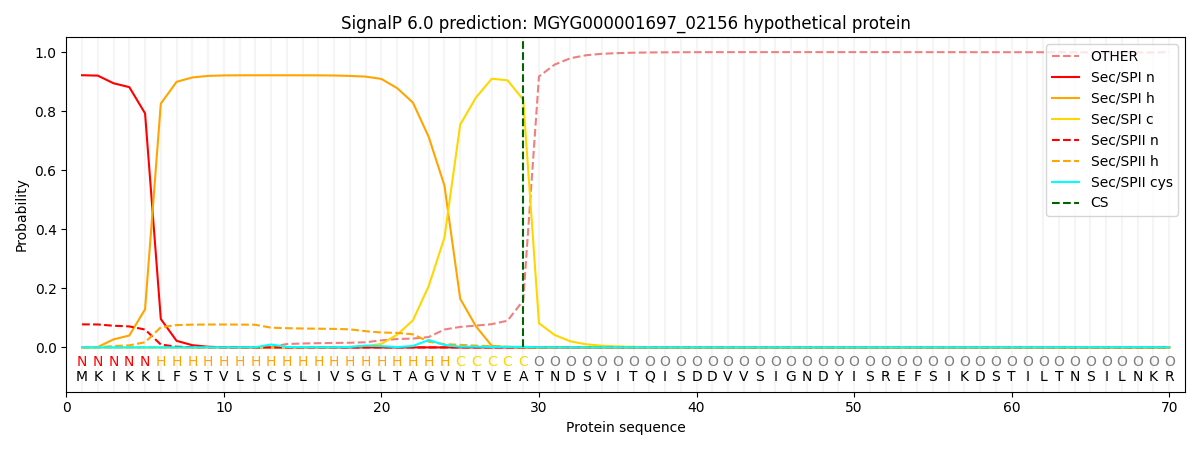
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001505 | 0.914260 | 0.083266 | 0.000366 | 0.000302 | 0.000282 |