You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001698_01923
You are here: Home > Sequence: MGYG000001698_01923
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Marvinbryantia formatexigens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Marvinbryantia; Marvinbryantia formatexigens | |||||||||||
| CAZyme ID | MGYG000001698_01923 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 129836; End: 133216 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 427 | 634 | 2.8e-24 | 0.8564814814814815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 3.57e-07 | 446 | 546 | 126 | 220 | beta-glucosidase BglX. |
| PRK07003 | PRK07003 | 0.002 | 94 | 236 | 425 | 560 | DNA polymerase III subunit gamma/tau. |
| PRK12372 | PRK12372 | 0.002 | 93 | 227 | 282 | 411 | ribonuclease III; Reviewed |
| PRK05035 | PRK05035 | 0.004 | 94 | 229 | 527 | 683 | electron transport complex protein RnfC; Provisional |
| PRK07003 | PRK07003 | 0.004 | 90 | 231 | 432 | 577 | DNA polymerase III subunit gamma/tau. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCJ97597.1 | 1.76e-222 | 288 | 1049 | 82 | 847 |
| QOY60939.1 | 1.25e-146 | 286 | 1046 | 57 | 809 |
| QIA06569.1 | 5.99e-145 | 281 | 1041 | 42 | 827 |
| QGY45729.1 | 2.47e-139 | 281 | 1041 | 42 | 832 |
| AGF59116.1 | 2.01e-138 | 285 | 1036 | 30 | 799 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z87_A | 4.29e-15 | 337 | 856 | 34 | 546 | ChainA, EmGH1 [Aurantiacibacter marinus],5Z87_B Chain B, EmGH1 [Aurantiacibacter marinus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46684 | 7.83e-44 | 316 | 753 | 35 | 496 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| B8NGU6 | 6.58e-42 | 332 | 1025 | 35 | 623 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
| Q2UFP8 | 3.98e-41 | 338 | 1025 | 45 | 627 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| Q5BCC6 | 2.41e-40 | 336 | 1006 | 35 | 603 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| P27034 | 1.59e-06 | 444 | 762 | 65 | 335 | Beta-glucosidase OS=Rhizobium radiobacter OX=358 GN=cbg-1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
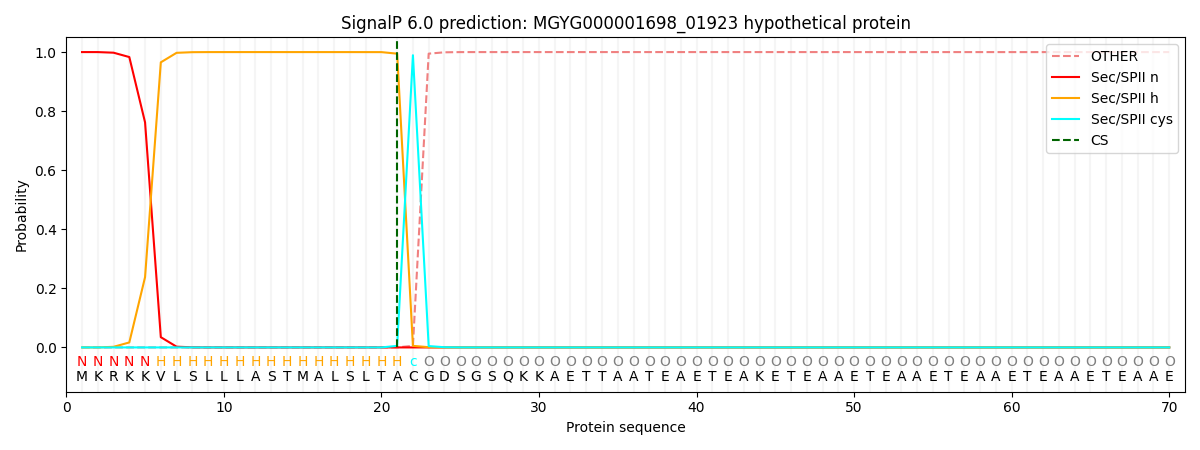
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000055 | 0.000000 | 0.000000 | 0.000000 |
