You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001698_02556
You are here: Home > Sequence: MGYG000001698_02556
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Marvinbryantia formatexigens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Marvinbryantia; Marvinbryantia formatexigens | |||||||||||
| CAZyme ID | MGYG000001698_02556 | |||||||||||
| CAZy Family | GH85 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35420; End: 36868 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH85 | 171 | 477 | 1.1e-100 | 0.9365079365079365 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4724 | COG4724 | 1.53e-142 | 112 | 476 | 27 | 374 | Endo-beta-N-acetylglucosaminidase D [Carbohydrate transport and metabolism]. |
| cd06547 | GH85_ENGase | 2.06e-114 | 166 | 476 | 1 | 288 | Endo-beta-N-acetylglucosaminidase (ENGase) hydrolyzes the N-N'-diacetylchitobiosyl core of N-glycosylproteins. The beta-1,4-glycosyl bond located between two N-acetylglucosamine residues is hydrolyzed such that N-acetylglucosamine 1 remains with the protein and N-acetylglucosamine 2 forms the reducing end of the released glycan. ENGase is a key enzyme in the processing of free oligosaccharides in the cytosol of eukaryotes. Oligosaccharides formed in the lumen of the endoplasmic reticulum are transported into the cytosol where they are catabolized by cytosolic ENGases and other enzymes, possibly to maximize the reutilization of the component sugars. ENGases have an eight-stranded alpha/beta barrel topology and are classified as a family 85 glycosyl hydrolase (GH85) domain. The GH85 ENGases are sequence-similar to the family 18 glycosyl hydrolases, also known as GH18 chitinases. An ENGase-like protein is also found in bacteria and is included in this alignment model. |
| pfam03644 | Glyco_hydro_85 | 1.03e-108 | 183 | 476 | 1 | 275 | Glycosyl hydrolase family 85. Family of endo-beta-N-acetylglucosaminidases. These enzymes work on a broad spectrum of substrates. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFZ80414.1 | 2.11e-195 | 91 | 476 | 39 | 423 |
| AUD76959.1 | 6.31e-195 | 91 | 476 | 29 | 413 |
| AHJ19483.1 | 6.31e-195 | 91 | 476 | 29 | 413 |
| AUD89384.1 | 6.31e-195 | 91 | 476 | 29 | 413 |
| AUD72994.1 | 6.31e-195 | 91 | 476 | 29 | 413 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VTF_A | 2.14e-102 | 114 | 476 | 11 | 354 | X-raycrystal structure of the Endo-beta-N-acetylglucosaminidase from Arthrobacter protophormiae E173Q mutant reveals a TIM barrel catalytic domain and two ancillary domains [Glutamicibacter protophormiae],2VTF_B X-ray crystal structure of the Endo-beta-N-acetylglucosaminidase from Arthrobacter protophormiae E173Q mutant reveals a TIM barrel catalytic domain and two ancillary domains [Glutamicibacter protophormiae] |
| 3FHA_A | 5.23e-102 | 114 | 476 | 6 | 349 | ChainA, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae],3FHA_B Chain B, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae],3FHA_C Chain C, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae],3FHA_D Chain D, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae] |
| 3FHQ_A | 5.23e-102 | 114 | 476 | 6 | 349 | ChainA, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae],3FHQ_B Chain B, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae],3FHQ_D Chain D, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae],3FHQ_F Chain F, Endo-beta-N-acetylglucosaminidase [Glutamicibacter protophormiae] |
| 2W91_A | 8.16e-54 | 122 | 476 | 18 | 355 | Structureof a Streptococcus pneumoniae family 85 glycoside hydrolase, Endo-D. [Streptococcus pneumoniae TIGR4],2W92_A Structure of a Streptococcus pneumoniae family 85 glycoside hydrolase, Endo-D, in complex with NAG-thiazoline. [Streptococcus pneumoniae TIGR4] |
| 3GDB_A | 2.82e-53 | 84 | 476 | 121 | 506 | Crystalstructure of Spr0440 glycoside hydrolase domain, Endo-D from Streptococcus pneumoniae R6 [Streptococcus pneumoniae R6] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1L251 | 1.94e-23 | 122 | 480 | 69 | 402 | Cytosolic endo-beta-N-acetylglucosaminidase OS=Danio rerio OX=7955 GN=engase PE=2 SV=1 |
| P0C7A1 | 1.12e-22 | 122 | 446 | 89 | 381 | Cytosolic endo-beta-N-acetylglucosaminidase OS=Gallus gallus OX=9031 GN=ENGASE PE=1 SV=1 |
| Q9SRL4 | 6.12e-22 | 194 | 446 | 99 | 327 | Cytosolic endo-beta-N-acetylglucosaminidase 2 OS=Arabidopsis thaliana OX=3702 GN=ENGASE2 PE=1 SV=1 |
| Q8BX80 | 1.52e-21 | 183 | 446 | 135 | 375 | Cytosolic endo-beta-N-acetylglucosaminidase OS=Mus musculus OX=10090 GN=Engase PE=1 SV=1 |
| Q8NFI3 | 1.53e-20 | 192 | 451 | 152 | 388 | Cytosolic endo-beta-N-acetylglucosaminidase OS=Homo sapiens OX=9606 GN=ENGASE PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
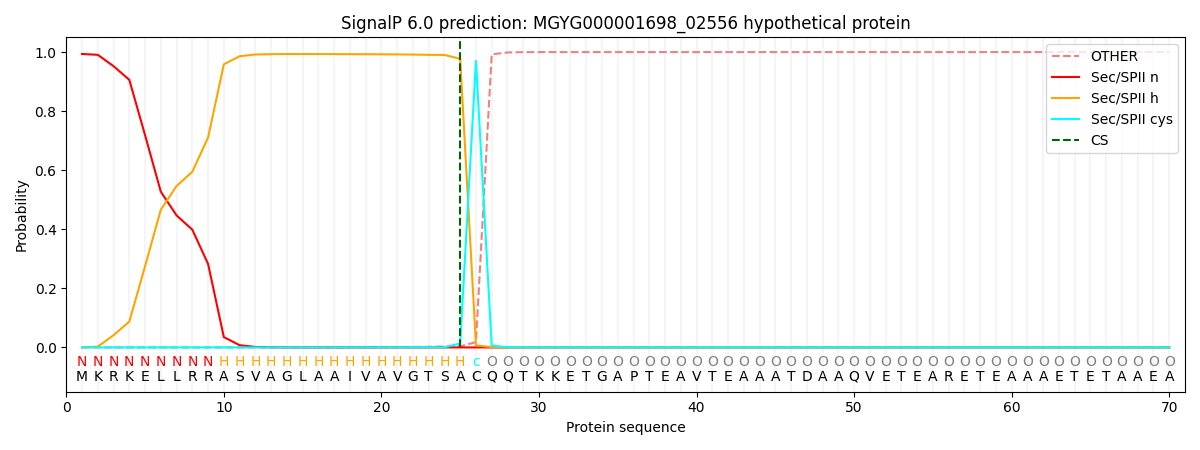
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000019 | 0.993660 | 0.000000 | 0.006346 | 0.000000 |
