You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001699_00659
You are here: Home > Sequence: MGYG000001699_00659
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
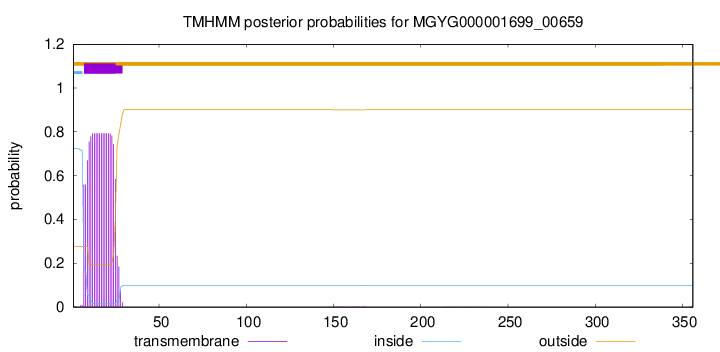
TMHMM annotations
Basic Information help
| Species | Clostridium_J ihumii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_J; Clostridium_J ihumii | |||||||||||
| CAZyme ID | MGYG000001699_00659 | |||||||||||
| CAZy Family | GH113 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 75618; End: 76688 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH113 | 52 | 333 | 2.5e-42 | 0.8594771241830066 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd19608 | GH113_mannanase-like | 1.20e-43 | 45 | 354 | 12 | 310 | Glycoside hydrolase family 113 beta-1,4-mannanase and similar proteins. Mannan endo-1,4-beta mannosidase (E.C 3.2.1.78) randomly cleaves (1->4)-beta-D-mannosidic linkages in mannans, galactomannans and glucomannans and is also called beta-1,4-mannanase, endo-1,4-beta-mannanase, endo-beta-1,4-mannase, beta-mannanase B, beta-1, 4-mannan 4-mannanohydrolase, endo-beta-mannanase, beta-D-mannanase, 1,4-beta-D-mannan mannanohydrolase, and 4-beta-D-mannan mannanohydrolase. (1->4)-beta-linked mannans are polysaccharides with a linear polymer backbone of (1->4)-beta-linked mannose units (in plants and fungi) or alternating mannose and glucose/galactose units (glucomannan in plants and fungi, and galactomannan and galactoglucomannan in plants), such as in the hemicellulose fraction of hard- and softwoods. Complete degradation of mannan requires a series of enzymes, including beta-1,4-mannanase. According to the CAZy database beta-1,4-mannanases are grouped into various glycoside hydrolase (GH) families; GH family 113 beta-1,4-mannanases include mostly bacterial and archaeal sequences. |
| cd19606 | GH113-like | 1.21e-10 | 161 | 330 | 137 | 271 | Glycoside hydrolase family 113 beta-mannosidase and similar proteins. Family 113 glycoside hydrolases cleave (1->4)-beta-glycosidic linkages, such as endo-1,4-beta-mannanase. This family also includes TIM-barrel domains found in gene transfer agent proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUN17378.1 | 1.16e-130 | 26 | 356 | 36 | 364 |
| AUM98762.1 | 1.16e-130 | 26 | 356 | 36 | 364 |
| APQ77895.1 | 1.16e-130 | 26 | 356 | 36 | 364 |
| CBZ03185.1 | 1.16e-130 | 26 | 356 | 36 | 364 |
| AXG95299.1 | 3.32e-130 | 26 | 356 | 36 | 364 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5YLH_A | 9.52e-06 | 161 | 329 | 140 | 273 | Structureof GH113 beta-1,4-mannanase [Amphibacillus xylanus NBRC 15112],5YLH_B Structure of GH113 beta-1,4-mannanase [Amphibacillus xylanus NBRC 15112] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
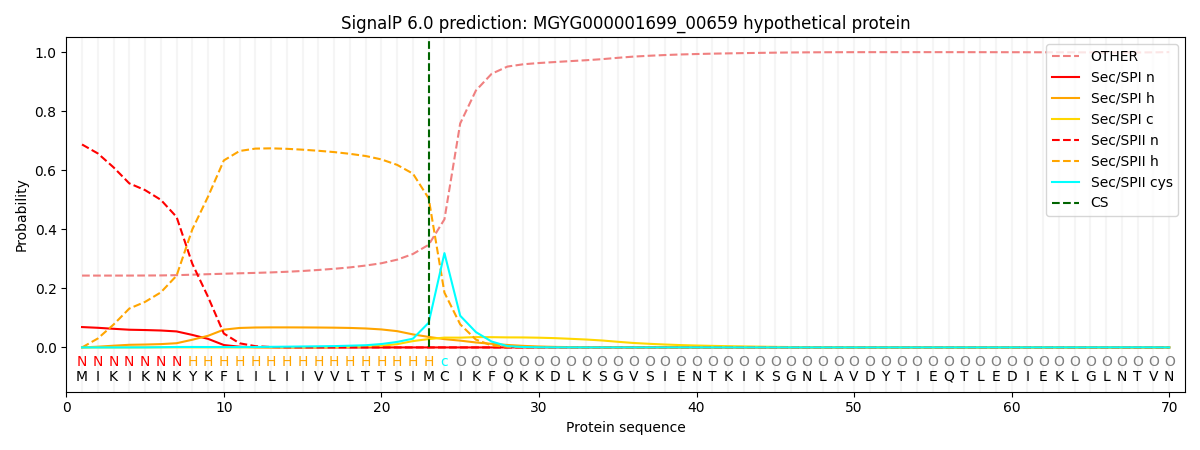
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.245456 | 0.064805 | 0.688442 | 0.000187 | 0.000173 | 0.000935 |