You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001701_02120
You are here: Home > Sequence: MGYG000001701_02120
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
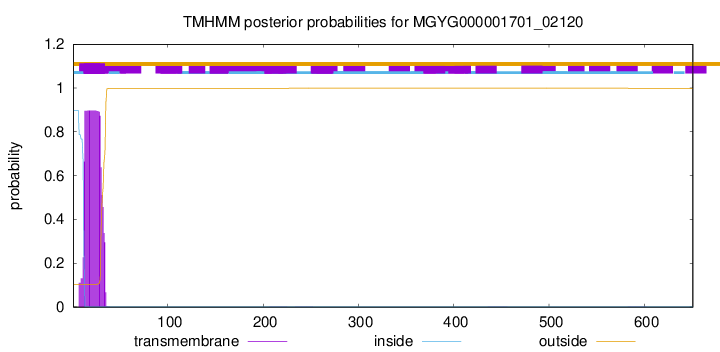
TMHMM annotations
Basic Information help
| Species | Corynebacterium afermentans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium afermentans | |||||||||||
| CAZyme ID | MGYG000001701_02120 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 97155; End: 99110 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 103 | 373 | 3.5e-53 | 0.9779735682819384 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0627 | FrmB | 3.85e-68 | 101 | 379 | 47 | 316 | S-formylglutathione hydrolase FrmB [Defense mechanisms]. |
| COG5479 | Psp3 | 3.68e-50 | 417 | 649 | 124 | 374 | Uncharacterized conserved protein, contains LGFP repeats [Function unknown]. |
| pfam00756 | Esterase | 5.16e-40 | 101 | 371 | 1 | 246 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
| pfam08310 | LGFP | 1.26e-20 | 457 | 508 | 1 | 52 | LGFP repeat. This 54 amino acid repeat is found in many hypothetical proteins. Several hypothetical proteins from C.glutamicum and C.efficiens along with PS1 protein contain this repeat region. The N-terminus region of PS1 contains an esterase domain which transfers corynomycolic acid. The C-terminus region consists of 4 tandem LGFP repeats. It is hypothesized that the PS1 proteins in Corynebacterium, when associated with the cell wall, may be anchored via the LGFP tandem repeats that may be important for maintaining cell wall integrity [Adindla et al. Comparative and Functional Genomics 2004; 5:2-16]. Deletion of Corynebacterium glutamicum csp1 protein results in a 10-fold increase in the cell volume of the organism and infers the corresponding proteins involvement in the cell shape formation. The secondary structure of each repeat is predicted to comprise two beta-strands and one alpha-helix [Adindla et al. 2004]. |
| pfam08310 | LGFP | 5.51e-16 | 563 | 613 | 1 | 52 | LGFP repeat. This 54 amino acid repeat is found in many hypothetical proteins. Several hypothetical proteins from C.glutamicum and C.efficiens along with PS1 protein contain this repeat region. The N-terminus region of PS1 contains an esterase domain which transfers corynomycolic acid. The C-terminus region consists of 4 tandem LGFP repeats. It is hypothesized that the PS1 proteins in Corynebacterium, when associated with the cell wall, may be anchored via the LGFP tandem repeats that may be important for maintaining cell wall integrity [Adindla et al. Comparative and Functional Genomics 2004; 5:2-16]. Deletion of Corynebacterium glutamicum csp1 protein results in a 10-fold increase in the cell volume of the organism and infers the corresponding proteins involvement in the cell shape formation. The secondary structure of each repeat is predicted to comprise two beta-strands and one alpha-helix [Adindla et al. 2004]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNP90090.1 | 0.0 | 1 | 650 | 1 | 646 |
| AIL97638.1 | 0.0 | 69 | 649 | 1 | 581 |
| AIJ34335.1 | 0.0 | 13 | 646 | 17 | 649 |
| SNV85099.1 | 0.0 | 13 | 646 | 17 | 649 |
| AOX04906.1 | 0.0 | 1 | 650 | 1 | 647 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6SX4_AAA | 2.01e-262 | 50 | 647 | 16 | 611 | ChainAAA, Protein PS1 [Corynebacterium glutamicum],6SX4_BBB Chain BBB, Protein PS1 [Corynebacterium glutamicum],6SX4_CCC Chain CCC, Protein PS1 [Corynebacterium glutamicum],6SX4_DDD Chain DDD, Protein PS1 [Corynebacterium glutamicum] |
| 6SWZ_AAA | 2.24e-94 | 401 | 647 | 7 | 249 | ChainAAA, Protein PS1 [Corynebacterium glutamicum] |
| 7MYG_A | 8.20e-48 | 130 | 375 | 30 | 276 | ChainA, Diacylglycerol acyltransferase [Mycobacterium tuberculosis],7MYG_B Chain B, Diacylglycerol acyltransferase [Mycobacterium tuberculosis] |
| 1DQZ_A | 8.67e-48 | 130 | 375 | 32 | 278 | ChainA, PROTEIN (ANTIGEN 85-C) [Mycobacterium tuberculosis],1DQZ_B Chain B, PROTEIN (ANTIGEN 85-C) [Mycobacterium tuberculosis] |
| 1DQY_A | 9.42e-48 | 130 | 375 | 35 | 281 | ChainA, PROTEIN (ANTIGEN 85-C) [Mycobacterium tuberculosis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1D6 | 3.24e-270 | 11 | 647 | 22 | 654 | Protein PS1 OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=csp1 PE=1 SV=1 |
| P0C1D7 | 6.11e-268 | 11 | 647 | 22 | 654 | Protein PS1 OS=Corynebacterium melassecola OX=41643 GN=csp1 PE=4 SV=1 |
| Q49575 | 1.30e-46 | 128 | 375 | 74 | 322 | Diacylglycerol acyltransferase/mycolyltransferase Ag85B OS=Mycobacterium intracellulare (strain ATCC 13950 / DSM 43223 / JCM 6384 / NCTC 13025 / 3600) OX=487521 GN=fbpB PE=3 SV=2 |
| P0A4V5 | 6.11e-46 | 130 | 375 | 80 | 326 | Diacylglycerol acyltransferase/mycolyltransferase Ag85C OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=fbpC PE=3 SV=1 |
| P9WQN9 | 6.11e-46 | 130 | 375 | 80 | 326 | Diacylglycerol acyltransferase/mycolyltransferase Ag85C OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=fbpC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
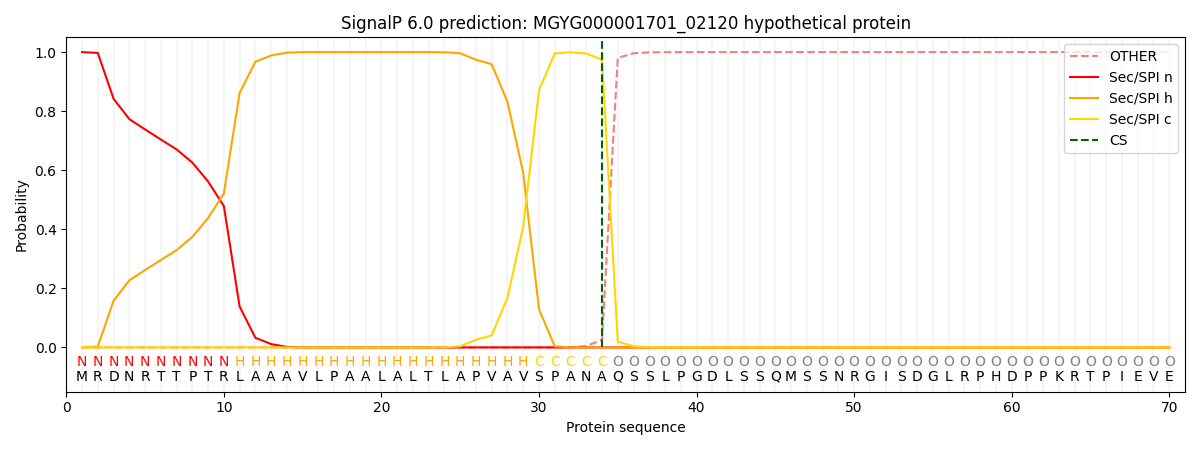
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000397 | 0.998783 | 0.000204 | 0.000224 | 0.000194 | 0.000165 |