You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001701_02122
Basic Information
help
| Species |
Corynebacterium afermentans
|
| Lineage |
Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium afermentans
|
| CAZyme ID |
MGYG000001701_02122
|
| CAZy Family |
GT89 |
| CAZyme Description |
Terminal beta-(1->2)-arabinofuranosyltransferase
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001701 |
2240769 |
Isolate |
France |
Europe |
|
| Gene Location |
Start: 100472;
End: 102283
Strand: -
|
| Family |
Start |
End |
Evalue |
family coverage |
| GT89 |
8 |
570 |
5.7e-195 |
0.9802158273381295 |
MGYG000001701_02122 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
Q8NLR0
|
7.03e-193 |
7 |
598 |
50 |
671 |
Terminal beta-(1->2)-arabinofuranosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=aftB PE=3 SV=1 |
|
O53582
|
1.19e-104 |
7 |
555 |
8 |
594 |
Terminal beta-(1->2)-arabinofuranosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=aftB PE=1 SV=3 |
This protein is predicted as OTHER
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.997704
|
0.000769
|
0.001154
|
0.000004
|
0.000003
|
0.000376
|
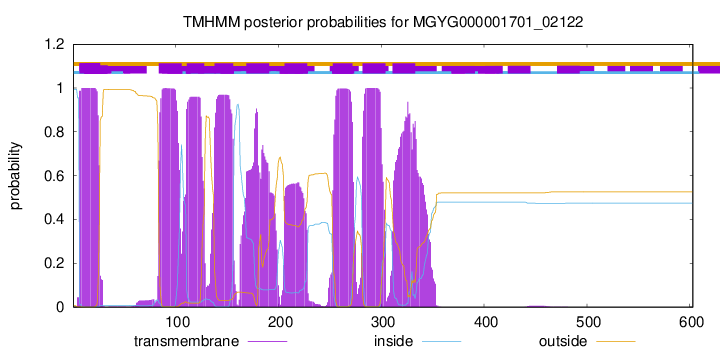
| start |
end |
| 7 |
26 |
| 84 |
106 |
| 111 |
128 |
| 138 |
156 |
| 169 |
191 |
| 206 |
228 |
| 253 |
272 |
| 282 |
304 |
| 311 |
333 |


