You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001702_03464
You are here: Home > Sequence: MGYG000001702_03464
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Virgibacillus picturae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_D; Amphibacillaceae; Virgibacillus; Virgibacillus picturae | |||||||||||
| CAZyme ID | MGYG000001702_03464 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43258; End: 45342 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 295 | 432 | 6.2e-20 | 0.9609375 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.68e-69 | 211 | 450 | 19 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| smart00047 | LYZ2 | 2.18e-28 | 283 | 440 | 1 | 146 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| pfam08239 | SH3_3 | 1.26e-12 | 480 | 531 | 1 | 54 | Bacterial SH3 domain. |
| NF033190 | inl_like_NEAT_1 | 5.45e-11 | 85 | 251 | 582 | 745 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam01832 | Glucosaminidase | 1.08e-09 | 295 | 384 | 1 | 76 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVR00509.1 | 0.0 | 1 | 694 | 13 | 708 |
| QRZ18453.1 | 6.93e-290 | 1 | 694 | 1 | 698 |
| API91764.1 | 7.41e-272 | 1 | 622 | 1 | 627 |
| AUS85359.1 | 1.55e-136 | 12 | 688 | 11 | 614 |
| QPQ32424.1 | 2.03e-136 | 12 | 694 | 11 | 621 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 1.14e-39 | 265 | 450 | 63 | 244 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI7_A | 4.55e-35 | 218 | 423 | 9 | 201 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 2.97e-34 | 218 | 423 | 9 | 201 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXP_A | 1.09e-33 | 242 | 434 | 49 | 230 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
| 6U0O_B | 1.22e-33 | 242 | 434 | 79 | 260 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 3.03e-52 | 242 | 450 | 682 | 880 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q5HQB9 | 5.12e-38 | 242 | 450 | 1131 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| O33635 | 5.12e-38 | 242 | 450 | 1131 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q8CPQ1 | 1.21e-37 | 242 | 450 | 1131 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| Q5HH31 | 1.12e-35 | 201 | 450 | 1007 | 1256 | Bifunctional autolysin OS=Staphylococcus aureus (strain COL) OX=93062 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
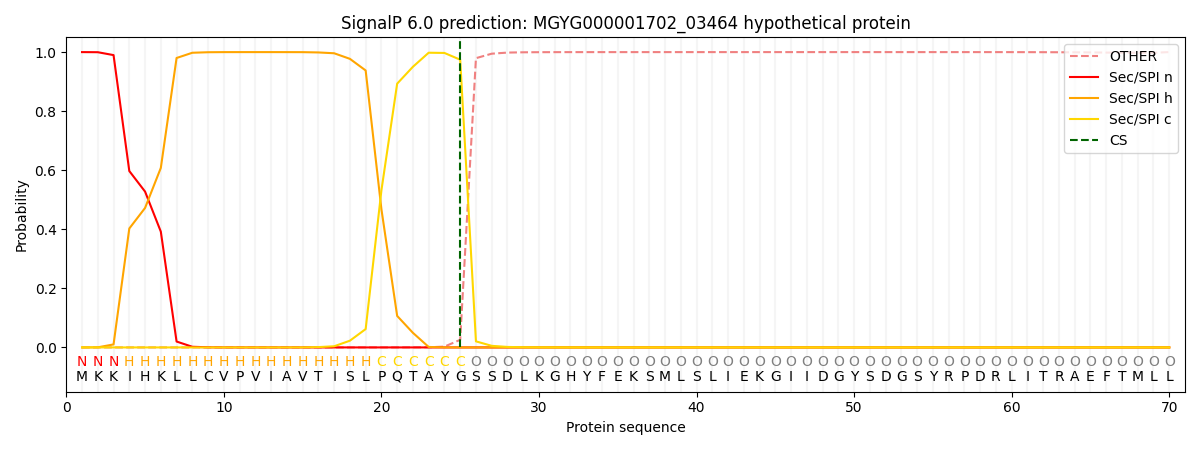
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004364 | 0.993818 | 0.000441 | 0.000476 | 0.000413 | 0.000476 |
