You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001705_03362
You are here: Home > Sequence: MGYG000001705_03362
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Citrobacter portucalensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Citrobacter; Citrobacter portucalensis | |||||||||||
| CAZyme ID | MGYG000001705_03362 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | Putative acyl-CoA thioester hydrolase YbhC | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3478991; End: 3480304 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10531 | PRK10531 | 0.0 | 12 | 430 | 2 | 422 | putative acyl-CoA thioester hydrolase. |
| COG4677 | PemB | 0.0 | 12 | 431 | 2 | 405 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| pfam01095 | Pectinesterase | 1.25e-10 | 91 | 429 | 1 | 266 | Pectinesterase. |
| PLN02488 | PLN02488 | 1.22e-08 | 91 | 437 | 198 | 482 | probable pectinesterase/pectinesterase inhibitor |
| PLN02708 | PLN02708 | 3.89e-08 | 79 | 316 | 216 | 413 | Probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBV41801.1 | 0.0 | 1 | 437 | 1 | 437 |
| BBW40138.1 | 0.0 | 1 | 437 | 1 | 437 |
| BBV46716.1 | 0.0 | 1 | 437 | 1 | 437 |
| BBV51990.1 | 0.0 | 1 | 437 | 1 | 437 |
| BBW12720.1 | 0.0 | 1 | 437 | 1 | 437 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GRH_A | 2.22e-252 | 39 | 437 | 24 | 422 | Crystalstructure of escherichia coli ybhc [Escherichia coli K-12] |
| 4PMH_A | 3.46e-65 | 43 | 429 | 1 | 366 | Thestructure of rice weevil pectin methyl esterase [Sitophilus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P46130 | 6.37e-269 | 11 | 437 | 1 | 427 | Putative acyl-CoA thioester hydrolase YbhC OS=Escherichia coli (strain K12) OX=83333 GN=ybhC PE=1 SV=2 |
| Q47474 | 2.86e-116 | 11 | 423 | 1 | 426 | Pectinesterase B OS=Dickeya dadantii (strain 3937) OX=198628 GN=pemB PE=1 SV=2 |
| P55743 | 6.83e-109 | 45 | 424 | 5 | 396 | Pectinesterase B OS=Pectobacterium parmentieri OX=1905730 GN=pemB PE=3 SV=2 |
| E7CIP7 | 2.93e-64 | 43 | 429 | 17 | 382 | Pectinesterase OS=Sitophilus oryzae OX=7048 GN=CE8-1 PE=1 SV=1 |
| P58601 | 1.22e-30 | 49 | 429 | 30 | 394 | Pectinesterase OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=pme PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
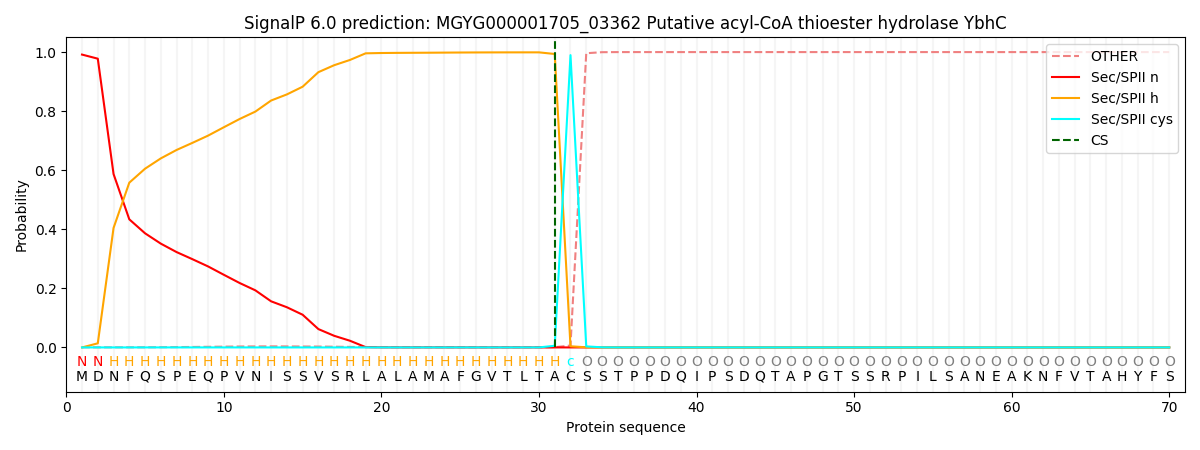
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000078 | 0.002046 | 0.997902 | 0.000013 | 0.000003 | 0.000001 |
