You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001713_02285
You are here: Home > Sequence: MGYG000001713_02285
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Vibrio furnissii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Vibrionaceae; Vibrio; Vibrio furnissii | |||||||||||
| CAZyme ID | MGYG000001713_02285 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 243653; End: 245656 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 67 | 349 | 2.3e-102 | 0.9896193771626297 |
| CBM27 | 501 | 662 | 1.9e-51 | 0.9880952380952381 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3934 | COG3934 | 2.50e-62 | 42 | 665 | 2 | 583 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| pfam09212 | CBM27 | 3.35e-31 | 497 | 664 | 7 | 172 | Carbohydrate binding module 27. Members of this family are carbohydrate binding modules that bind to beta-1, 4-manno-oligosaccharides, carob galactomannan, and konjac glucomannan, but not to cellulose (insoluble and soluble) or soluble birchwood xylan. They adopt a beta sandwich structure comprising 13 beta strands with a single, small alpha-helix and a single metal atom. |
| pfam00150 | Cellulase | 2.63e-09 | 48 | 327 | 5 | 247 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 0.003 | 25 | 207 | 36 | 192 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDC94579.1 | 0.0 | 1 | 667 | 1 | 667 |
| AMF93025.1 | 0.0 | 1 | 667 | 1 | 667 |
| ADT88760.1 | 0.0 | 1 | 667 | 1 | 667 |
| BAG69482.2 | 0.0 | 1 | 667 | 1 | 669 |
| QIL87027.1 | 0.0 | 1 | 667 | 1 | 667 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6TN6_A | 4.07e-171 | 45 | 661 | 7 | 636 | X-raystructure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
| 3PZ9_A | 6.07e-134 | 43 | 386 | 19 | 381 | Nativestructure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
| 4QP0_A | 1.08e-81 | 34 | 388 | 2 | 356 | CrystalStructure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
| 4AWE_A | 4.04e-62 | 36 | 389 | 6 | 382 | TheCrystal Structure of Chrysonilia sitophila endo-beta-D-1,4- mannanase [Neurospora sitophila] |
| 3ZIZ_A | 1.87e-60 | 41 | 391 | 20 | 357 | ChainA, Gh5 Endo-beta-1,4-mannanase [Podospora anserina] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1D8Y6 | 4.02e-62 | 32 | 371 | 22 | 358 | Probable mannan endo-1,4-beta-mannosidase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=manA PE=3 SV=1 |
| Q5AZ53 | 1.57e-61 | 36 | 388 | 22 | 392 | Mannan endo-1,4-beta-mannosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manC PE=1 SV=1 |
| B0Y9E7 | 2.20e-61 | 23 | 362 | 85 | 421 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=manF PE=3 SV=2 |
| Q4WBS1 | 2.20e-61 | 23 | 362 | 85 | 421 | Mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=manF PE=1 SV=2 |
| B8NVK8 | 3.04e-60 | 18 | 371 | 25 | 374 | Probable mannan endo-1,4-beta-mannosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
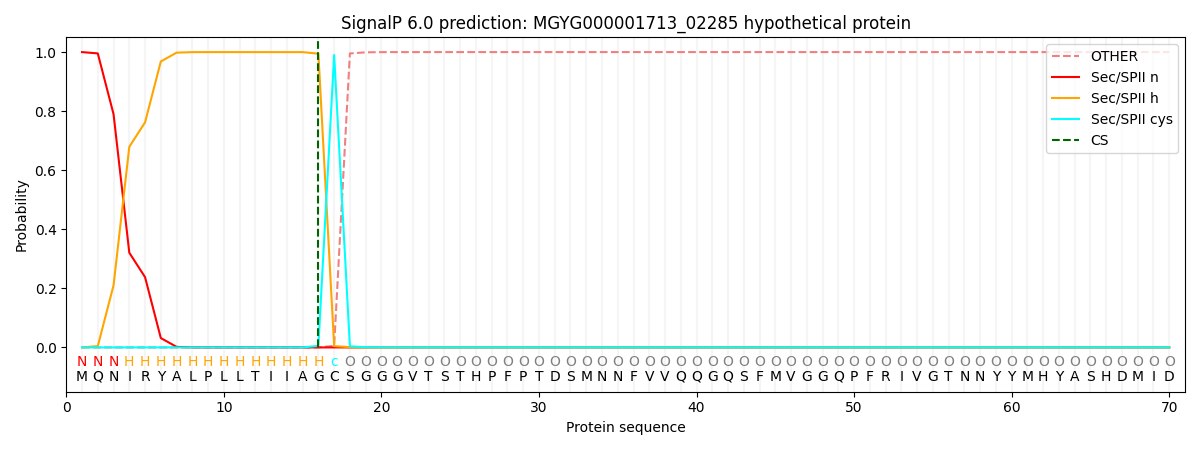
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000051 | 0.000000 | 0.000000 | 0.000000 |
