You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001720_00025
You are here: Home > Sequence: MGYG000001720_00025
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp002350355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp002350355 | |||||||||||
| CAZyme ID | MGYG000001720_00025 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | Mannan endo-1,4-beta-mannosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 31474; End: 32529 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 19 | 336 | 3e-65 | 0.9471947194719472 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 4.10e-36 | 18 | 243 | 5 | 230 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 2.29e-19 | 116 | 341 | 123 | 346 | Beta-mannanase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGH13968.1 | 1.53e-147 | 20 | 351 | 25 | 357 |
| BCS86241.1 | 5.50e-144 | 11 | 349 | 13 | 351 |
| QUB85259.1 | 6.46e-130 | 18 | 347 | 12 | 347 |
| QUB69107.1 | 5.01e-126 | 16 | 333 | 22 | 342 |
| QNT67784.1 | 2.24e-125 | 16 | 351 | 25 | 367 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ZXO_A | 1.97e-103 | 25 | 346 | 15 | 338 | Thestructure of a GH26 beta-mannanase from Bacteroides ovatus, BoMan26A. [Bacteroides ovatus] |
| 2VX4_A | 1.09e-70 | 18 | 342 | 38 | 386 | CellvibrioJaponicus Mannanase Cjman26c Native Form [Cellvibrio japonicus],2VX6_A CELLVIBRIO JAPONICUS MANNANASE CJMAN26C Gal1Man4-BOUND FORM [Cellvibrio japonicus] |
| 2VX5_A | 1.09e-70 | 18 | 342 | 38 | 386 | CellvibrioJaponicus Mannanase Cjman26c Mannose-Bound Form [Cellvibrio japonicus] |
| 4CD4_A | 2.02e-70 | 18 | 342 | 61 | 409 | Thestructure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManIFG [Cellvibrio japonicus Ueda107],4CD5_A The structure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManMIm [Cellvibrio japonicus Ueda107] |
| 2VX7_A | 8.55e-70 | 18 | 342 | 38 | 386 | CellvibrioJaponicus Mannanase Cjman26c Mannobiose-Bound Form [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49424 | 1.13e-37 | 18 | 312 | 54 | 363 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
| A1A278 | 7.64e-36 | 50 | 342 | 77 | 393 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| O05512 | 5.00e-23 | 47 | 316 | 66 | 331 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
| P49425 | 3.68e-22 | 32 | 281 | 163 | 409 | Mannan endo-1,4-beta-mannosidase OS=Rhodothermus marinus (strain ATCC 43812 / DSM 4252 / R-10) OX=518766 GN=manA PE=1 SV=3 |
| P55278 | 1.41e-20 | 60 | 243 | 77 | 266 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
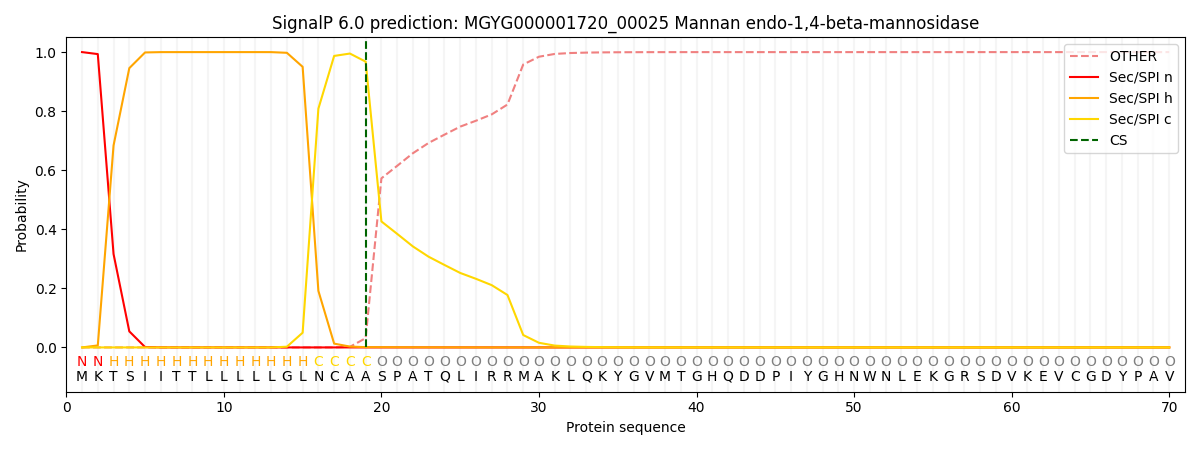
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000239 | 0.999229 | 0.000131 | 0.000139 | 0.000124 | 0.000120 |
