You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001720_01154
Basic Information
help
| Species |
Prevotella sp002350355
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp002350355
|
| CAZyme ID |
MGYG000001720_01154
|
| CAZy Family |
GH5 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 690 |
|
77171.71 |
6.3952 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001720 |
3435079 |
MAG |
Sweden |
Europe |
|
| Gene Location |
Start: 13895;
End: 15967
Strand: -
|
No EC number prediction in MGYG000001720_01154.
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
47 |
314 |
4.5e-116 |
0.9845559845559846 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam00150
|
Cellulase |
3.36e-11 |
146 |
307 |
72 |
235 |
Cellulase (glycosyl hydrolase family 5). |
| smart00635
|
BID_2 |
0.002 |
444 |
501 |
22 |
81 |
Bacterial Ig-like domain 2. |
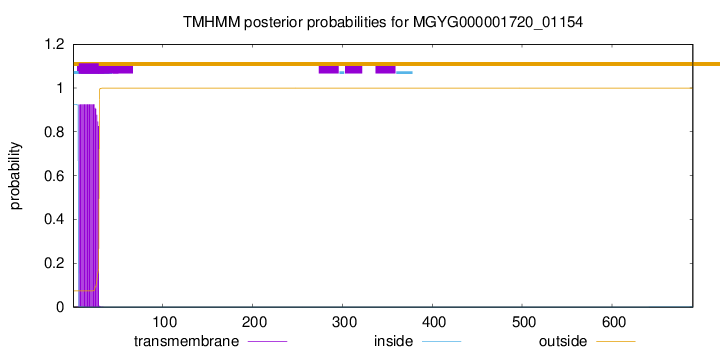
This protein is predicted as SP
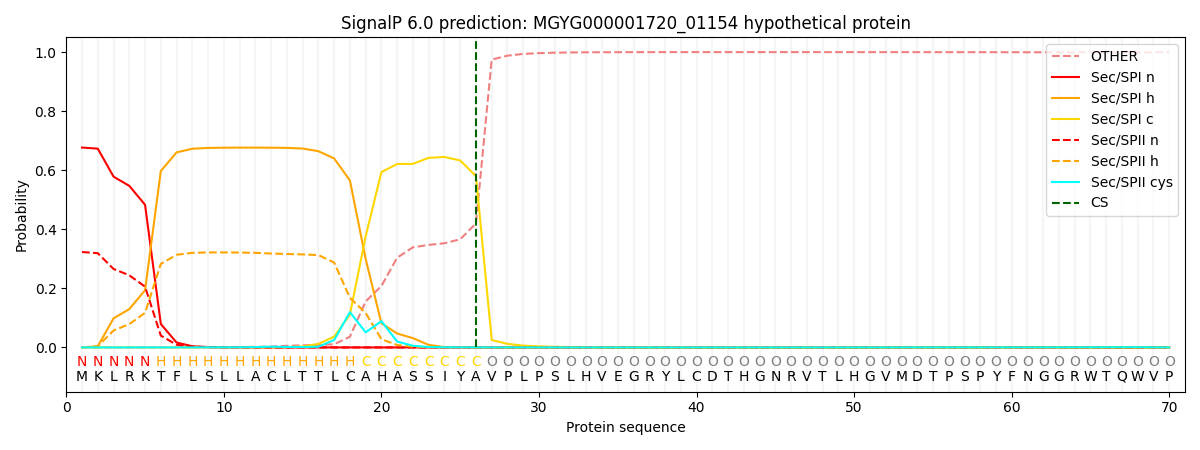
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000544
|
0.670867
|
0.327780
|
0.000370
|
0.000242
|
0.000188
|