You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001720_02088
You are here: Home > Sequence: MGYG000001720_02088
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp002350355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp002350355 | |||||||||||
| CAZyme ID | MGYG000001720_02088 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16699; End: 18447 Strand: - | |||||||||||
Full Sequence Download help
| MKQILFSFLA LSTVLSASAQ STYTIGFNPS EHYQTIMDFG ASDCWTADFV GRYFATTEKE | 60 |
| KAARLLFSQK MDSNGNPEGI GLSVWRVNLG AGSSEQGDGS NITDVTRRAD CYIKSDGTYD | 120 |
| WNRCAGQRWF MEQAKSYGVD HFLLFANSAP IYYTANGLAN NKDGAAGANL KEDCYDDFAS | 180 |
| YIATTARHFT DLGYPITYVD PVNEPAFDWR EGQEGSPWQN AEISRLVKEL DQALDKQQLS | 240 |
| GTSIVMPEAS AWDRLYQHCS DYGGRASDQI EAFWNPANSE TYIGGLGHVE KAVAGHDYWT | 300 |
| FGSDATLVDT RKKVAQKAAQ YGLKVMQTEW SMLDKEPDSD TGFPGSYDEA TDMDIALFMG | 360 |
| KLIYVGLTEG NMSSWSYWTA MAQSQWSQKN RFELLRLNAT GDTGYESYGD LTKGGTVTAN | 420 |
| PNLWVLGNYS RFVRPGYQRI AMTGTTVDDI DGLMATAFVS PDGKRIVAVF VNNGGAAKGV | 480 |
| KFNTSATAVH KYVTDATHSL SLDATLSTTP NADTRILIPA RSVVTFTLDG DFTTGIQSIE | 540 |
| EDAKKVNAVF SLTGMKVADD VSLLHSLPKG IYVVDGKKVV NH | 582 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 25 | 526 | 7e-166 | 0.9917355371900827 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14587 | Glyco_hydr_30_2 | 1.34e-116 | 24 | 380 | 3 | 355 | O-Glycosyl hydrolase family 30. |
| COG5520 | XynC | 9.26e-18 | 6 | 529 | 15 | 429 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam17189 | Glyco_hydro_30C | 9.70e-06 | 436 | 526 | 1 | 63 | Glycosyl hydrolase family 30 beta sandwich domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCS84534.1 | 2.49e-266 | 1 | 580 | 1 | 581 |
| VEH15029.1 | 2.61e-258 | 9 | 579 | 9 | 579 |
| EFI72736.1 | 2.85e-226 | 1 | 582 | 1 | 582 |
| QNT66374.1 | 3.09e-216 | 10 | 580 | 12 | 582 |
| QVJ80556.1 | 1.53e-215 | 30 | 580 | 197 | 732 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CLW_A | 3.61e-74 | 33 | 529 | 16 | 497 | Crystalstructure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_B Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_C Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_D Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_E Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_F Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 6M5Z_A | 1.59e-21 | 10 | 528 | 10 | 453 | ChainA, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612],6M5Z_B Chain B, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612] |
| 7NCX_AAA | 5.35e-14 | 20 | 528 | 7 | 454 | ChainAAA, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
| 7O0E_A | 1.98e-13 | 24 | 528 | 4 | 447 | ChainA, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464],7O0E_G Chain G, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
| 4FMV_A | 7.87e-10 | 161 | 526 | 99 | 384 | CrystalStructure Analysis of a GH30 Endoxylanase from Clostridium papyrosolvens C71 [Ruminiclostridium papyrosolvens DSM 2782] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G2Q1N4 | 2.25e-14 | 1 | 528 | 1 | 471 | GH30 family xylanase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Xyn30A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
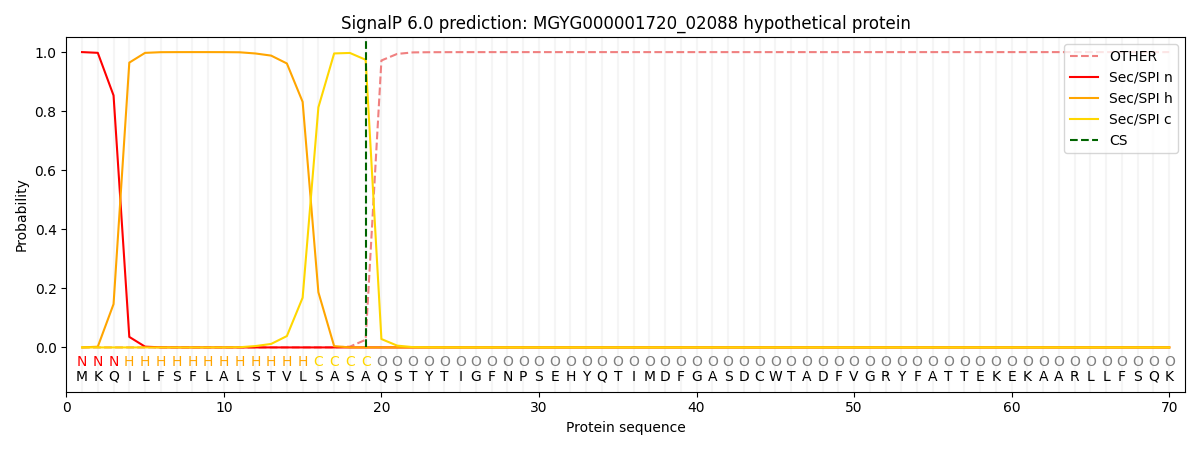
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000332 | 0.998965 | 0.000176 | 0.000182 | 0.000163 | 0.000151 |
