You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001720_02318
Basic Information
help
| Species |
Prevotella sp002350355
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp002350355
|
| CAZyme ID |
MGYG000001720_02318
|
| CAZy Family |
GH2 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001720 |
3435079 |
MAG |
Sweden |
Europe |
|
| Gene Location |
Start: 19676;
End: 22399
Strand: -
|
No EC number prediction in MGYG000001720_02318.
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| PRK10150
|
PRK10150 |
8.10e-04 |
46 |
167 |
11 |
136 |
beta-D-glucuronidase; Provisional |
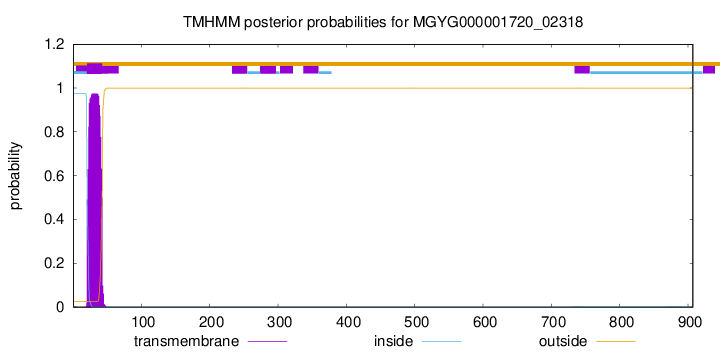
This protein is predicted as SP
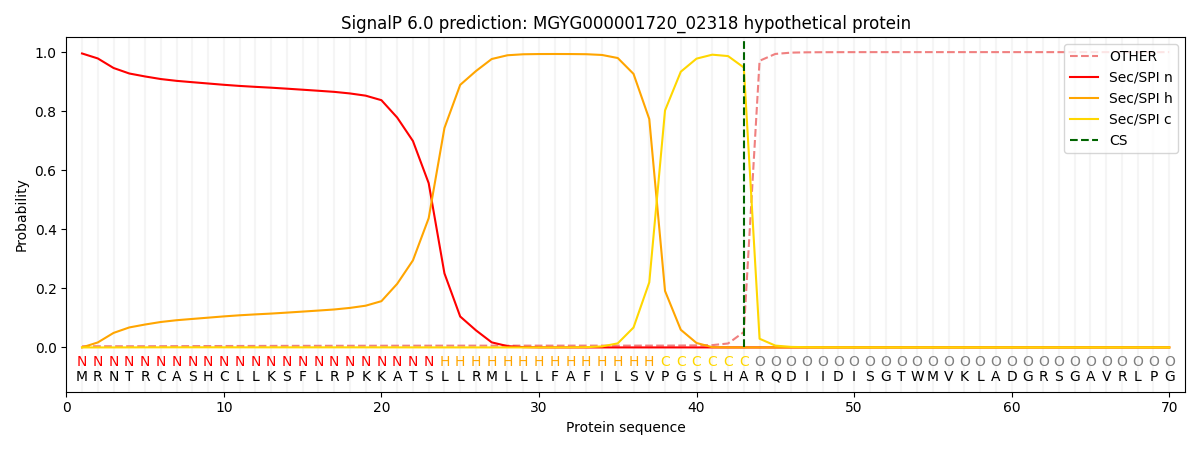
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.005210
|
0.992868
|
0.001264
|
0.000243
|
0.000196
|
0.000189
|