You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001732_00003
You are here: Home > Sequence: MGYG000001732_00003
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | HGM11530 sp900751685 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; HGM11530; HGM11530 sp900751685 | |||||||||||
| CAZyme ID | MGYG000001732_00003 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | Pectate lyase L | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6021; End: 7538 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 23 | 405 | 1.7e-91 | 0.9705882352941176 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWV81016.1 | 1.97e-164 | 13 | 408 | 19 | 412 |
| ADZ22427.1 | 1.97e-164 | 13 | 408 | 19 | 412 |
| AAK81317.1 | 1.97e-164 | 13 | 408 | 19 | 412 |
| AEI32814.1 | 1.97e-164 | 13 | 408 | 19 | 412 |
| AFH63317.1 | 1.08e-160 | 14 | 411 | 27 | 431 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLQ_A | 7.82e-57 | 26 | 409 | 3 | 428 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
| 1RU4_A | 5.55e-26 | 26 | 406 | 15 | 379 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 3VST_A | 9.19e-06 | 27 | 108 | 2 | 77 | Thecomplex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_B The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_C The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_D The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_A The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_B The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_C The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_D The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_A The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_B The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_C The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_D The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A6 | 2.24e-25 | 26 | 406 | 40 | 404 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 4.11e-25 | 26 | 406 | 40 | 404 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
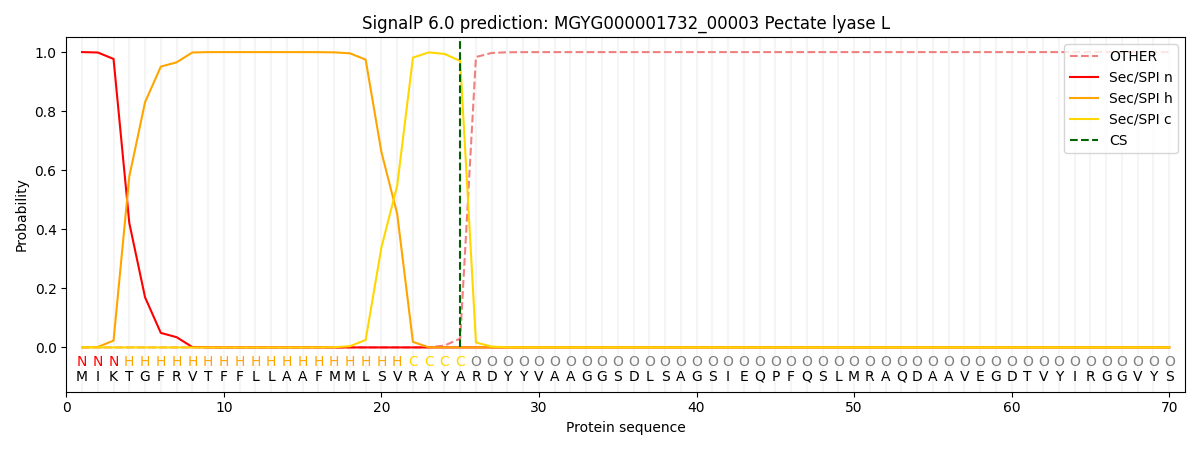
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000368 | 0.998827 | 0.000244 | 0.000186 | 0.000187 | 0.000178 |
