You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001732_00014
You are here: Home > Sequence: MGYG000001732_00014
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | HGM11530 sp900751685 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; HGM11530; HGM11530 sp900751685 | |||||||||||
| CAZyme ID | MGYG000001732_00014 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26210; End: 29533 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 43 | 496 | 1.3e-133 | 0.9824561403508771 |
| CBM62 | 742 | 861 | 2.1e-20 | 0.9007633587786259 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14587 | Glyco_hydr_30_2 | 1.02e-21 | 50 | 349 | 18 | 357 | O-Glycosyl hydrolase family 30. |
| COG5520 | XynC | 2.65e-14 | 134 | 445 | 102 | 403 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam00754 | F5_F8_type_C | 2.10e-09 | 742 | 861 | 2 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam02368 | Big_2 | 3.33e-06 | 668 | 730 | 11 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| pfam17132 | Glyco_hydro_106 | 2.21e-05 | 758 | 941 | 189 | 359 | alpha-L-rhamnosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADU23821.1 | 6.75e-145 | 32 | 857 | 34 | 774 |
| QTE67528.1 | 2.33e-136 | 24 | 498 | 25 | 510 |
| AOZ97766.1 | 7.37e-124 | 32 | 499 | 61 | 582 |
| ADL35667.1 | 2.90e-122 | 32 | 498 | 89 | 606 |
| CBK73885.1 | 3.44e-116 | 30 | 499 | 948 | 1476 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CLW_A | 5.49e-11 | 46 | 251 | 18 | 220 | Crystalstructure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_B Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_C Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_D Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_E Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_F Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 2YB7_A | 4.43e-07 | 756 | 860 | 27 | 133 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YB7_B Chain B, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YFU_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YFZ_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YG0_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 5NGK_A | 8.53e-07 | 22 | 418 | 58 | 437 | Theendo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGK_B The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGK_C The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_A The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_B The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_C The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron] |
| 5G56_A | 8.60e-07 | 665 | 860 | 643 | 834 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A401ETL2 | 1.19e-32 | 26 | 450 | 31 | 567 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
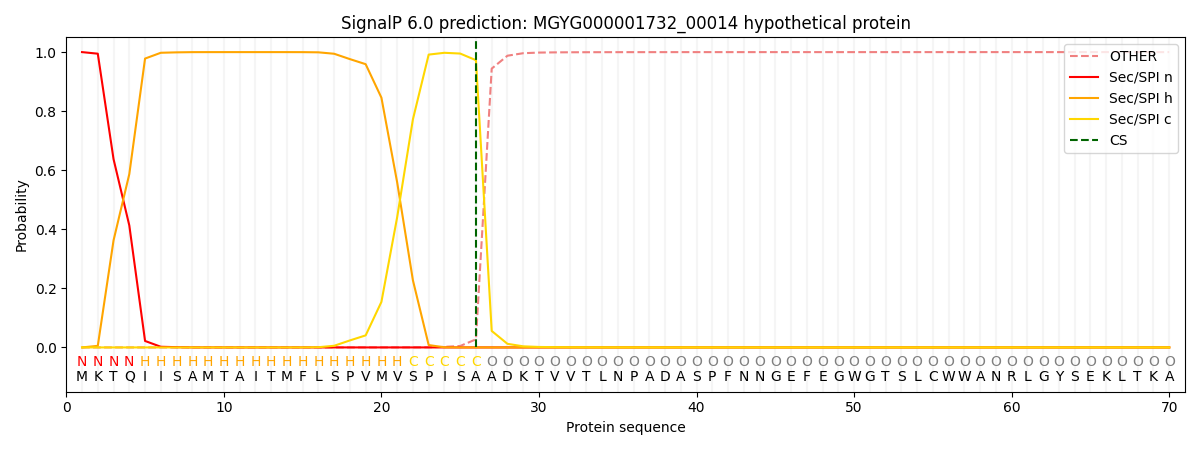
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000260 | 0.999100 | 0.000153 | 0.000178 | 0.000146 | 0.000134 |
