You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001735_01150
You are here: Home > Sequence: MGYG000001735_01150
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; | |||||||||||
| CAZyme ID | MGYG000001735_01150 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 129124; End: 130509 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 68 | 249 | 6.7e-16 | 0.44110275689223055 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 4.09e-34 | 67 | 418 | 1 | 335 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 4.23e-11 | 70 | 202 | 1 | 119 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| TIGR04380 | myo_inos_iolG | 1.23e-10 | 69 | 226 | 1 | 145 | inositol 2-dehydrogenase. All members of the seed alignment for this model are known or predicted inositol 2-dehydrogenase sequences co-clustered with other enzymes for catabolism of myo-inositol or closely related compounds. Inositol 2-dehydrogenase catalyzes the first step in inositol catabolism. Members of this family may vary somewhat in their ranges of acceptable substrates and some may act on analogs to myo-inositol rather than myo-inositol per se. [Energy metabolism, Sugars] |
| pfam10518 | TAT_signal | 4.04e-05 | 6 | 31 | 1 | 26 | TAT (twin-arginine translocation) pathway signal sequence. |
| TIGR01299 | synapt_SV2 | 5.86e-04 | 281 | 319 | 467 | 506 | synaptic vesicle protein SV2. This model describes a tightly conserved subfamily of the larger family of sugar (and other) transporters described by pfam00083. Members of this subfamily include closely related forms SV2A and SV2B of synaptic vesicle protein from vertebrates and a more distantly related homolog (below trusted cutoff) from Drosophila melanogaster. Members are predicted to have two sets of six transmembrane helices. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKH96209.1 | 0.0 | 1 | 460 | 1 | 460 |
| BBK93797.1 | 1.10e-317 | 1 | 460 | 1 | 460 |
| QIX65768.1 | 1.10e-317 | 1 | 460 | 1 | 460 |
| QUT96603.1 | 1.10e-317 | 1 | 460 | 1 | 460 |
| AST52349.1 | 1.10e-317 | 1 | 460 | 1 | 460 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CEA_A | 9.97e-09 | 70 | 295 | 9 | 210 | ChainA, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_B Chain B, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_C Chain C, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_D Chain D, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1] |
| 3E18_A | 9.70e-06 | 74 | 360 | 10 | 260 | CRYSTALSTRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua [Listeria innocua],3E18_B CRYSTAL STRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua [Listeria innocua] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as TAT
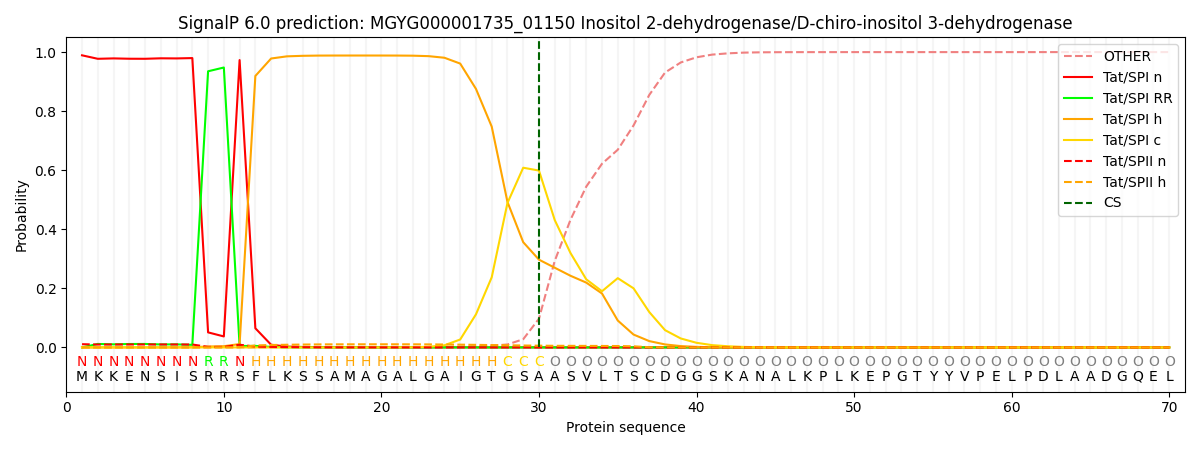
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 0.989101 | 0.010891 | 0.000000 |
