You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001738_00232
You are here: Home > Sequence: MGYG000001738_00232
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Duncaniella sp001701225 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Duncaniella; Duncaniella sp001701225 | |||||||||||
| CAZyme ID | MGYG000001738_00232 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5548; End: 7029 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 199 | 455 | 1.3e-43 | 0.8993055555555556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 1.94e-17 | 156 | 425 | 42 | 349 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02671 | PLN02671 | 7.14e-16 | 195 | 470 | 57 | 337 | pectinesterase |
| pfam01095 | Pectinesterase | 1.10e-14 | 200 | 433 | 3 | 243 | Pectinesterase. |
| PLN02773 | PLN02773 | 9.81e-14 | 193 | 486 | 1 | 299 | pectinesterase |
| PLN02432 | PLN02432 | 1.86e-12 | 199 | 470 | 13 | 272 | putative pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD38260.1 | 1.12e-249 | 18 | 493 | 19 | 496 |
| QCP71948.1 | 1.12e-249 | 18 | 493 | 19 | 496 |
| QBI04917.1 | 6.19e-157 | 22 | 493 | 10 | 490 |
| QBE65504.1 | 1.96e-155 | 31 | 493 | 37 | 507 |
| QJE01968.1 | 1.27e-150 | 4 | 493 | 10 | 508 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
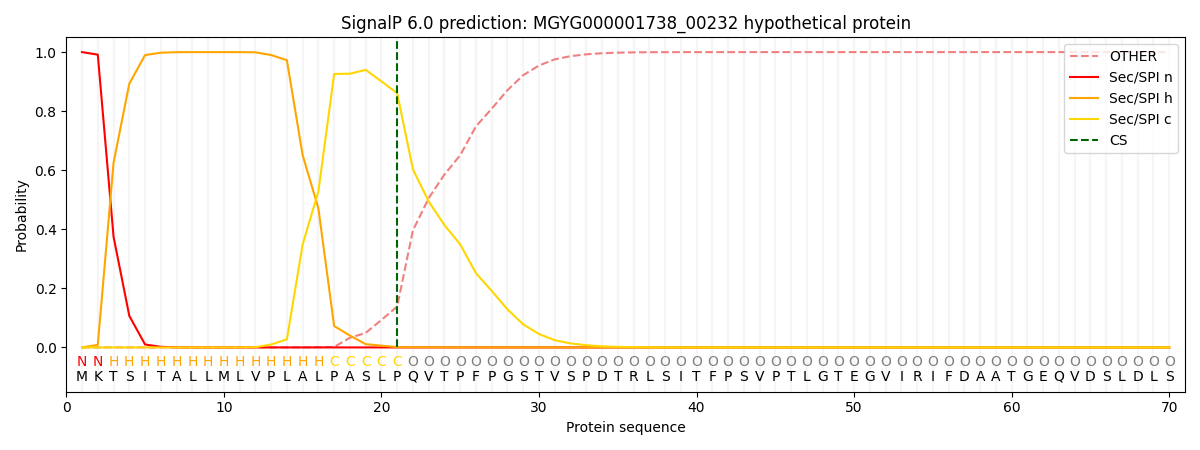
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000466 | 0.998810 | 0.000186 | 0.000181 | 0.000159 | 0.000158 |
