You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001738_00457
You are here: Home > Sequence: MGYG000001738_00457
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Duncaniella sp001701225 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Duncaniella; Duncaniella sp001701225 | |||||||||||
| CAZyme ID | MGYG000001738_00457 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 126687; End: 129692 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 619 | 938 | 4e-93 | 0.976897689768977 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 9.90e-107 | 617 | 937 | 1 | 308 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 1.51e-96 | 662 | 935 | 1 | 261 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.03e-74 | 616 | 935 | 22 | 335 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADX05746.1 | 1.62e-96 | 674 | 954 | 7 | 289 |
| QBI56661.1 | 1.12e-72 | 615 | 941 | 47 | 359 |
| ADX05712.1 | 2.76e-72 | 615 | 916 | 70 | 380 |
| QIJ64256.1 | 2.73e-69 | 616 | 937 | 37 | 342 |
| AIS00037.1 | 3.56e-69 | 616 | 937 | 37 | 342 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1XYZ_A | 3.50e-72 | 617 | 934 | 28 | 337 | ChainA, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus],1XYZ_B Chain B, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus] |
| 6FHE_A | 1.34e-65 | 628 | 937 | 23 | 338 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 1V6Y_A | 2.43e-62 | 621 | 934 | 9 | 307 | CrystalStructure Of chimeric Xylanase between Streptomyces Olivaceoviridis E-86 FXYN and Cellulomonas fimi Cex [Streptomyces olivaceoviridis] |
| 3W24_A | 2.30e-61 | 628 | 937 | 19 | 323 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 3W25_A | 1.53e-60 | 628 | 937 | 19 | 323 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W26_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylotriose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10478 | 9.23e-67 | 617 | 934 | 518 | 827 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| Q0H904 | 2.68e-58 | 648 | 941 | 59 | 325 | Endo-1,4-beta-xylanase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=xlnC PE=2 SV=2 |
| P23360 | 4.12e-58 | 629 | 934 | 45 | 320 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
| Q0CBM8 | 9.72e-58 | 646 | 934 | 58 | 319 | Probable endo-1,4-beta-xylanase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=xlnC PE=2 SV=2 |
| Q4JHP5 | 1.82e-57 | 646 | 934 | 58 | 319 | Probable endo-1,4-beta-xylanase C OS=Aspergillus terreus OX=33178 GN=xlnC PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
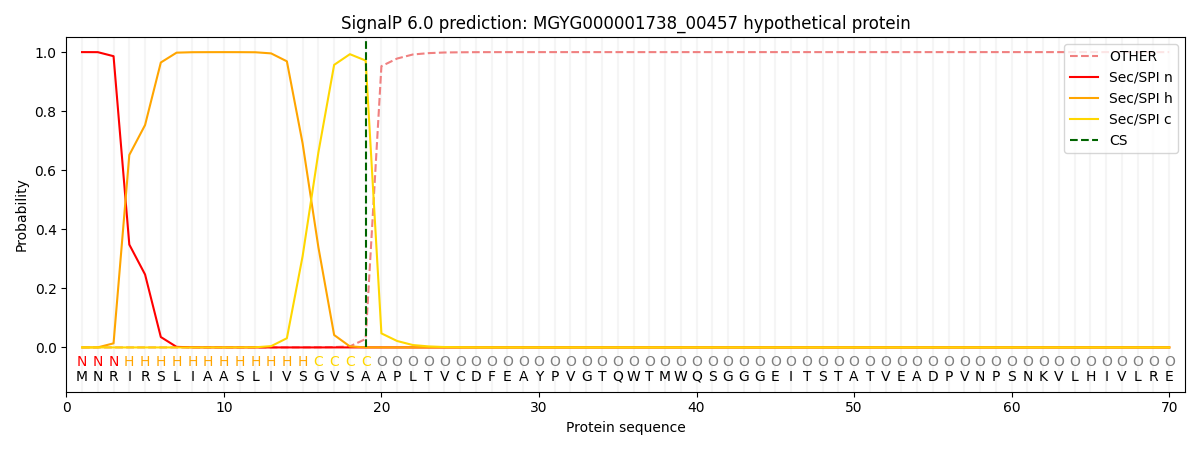
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000432 | 0.998744 | 0.000248 | 0.000205 | 0.000182 | 0.000161 |
