You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001738_01916
You are here: Home > Sequence: MGYG000001738_01916
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Duncaniella sp001701225 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Duncaniella; Duncaniella sp001701225 | |||||||||||
| CAZyme ID | MGYG000001738_01916 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16047; End: 18548 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 344 | 733 | 1.3e-88 | 0.49271844660194175 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 4.21e-139 | 32 | 809 | 2 | 863 | alpha-L-rhamnosidase. |
| pfam00754 | F5_F8_type_C | 0.008 | 186 | 298 | 30 | 120 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ47717.1 | 6.49e-225 | 4 | 830 | 3 | 918 |
| QRQ54617.1 | 6.49e-225 | 4 | 830 | 3 | 918 |
| EDO11490.1 | 6.49e-225 | 4 | 830 | 3 | 918 |
| SCV07115.1 | 6.49e-225 | 4 | 830 | 3 | 918 |
| AII65174.1 | 1.87e-218 | 21 | 831 | 37 | 927 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQM_A | 7.44e-80 | 15 | 830 | 19 | 919 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 1.90e-79 | 15 | 830 | 19 | 919 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 6Q2F_A | 4.51e-61 | 25 | 809 | 41 | 935 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 5.45e-62 | 22 | 833 | 26 | 740 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
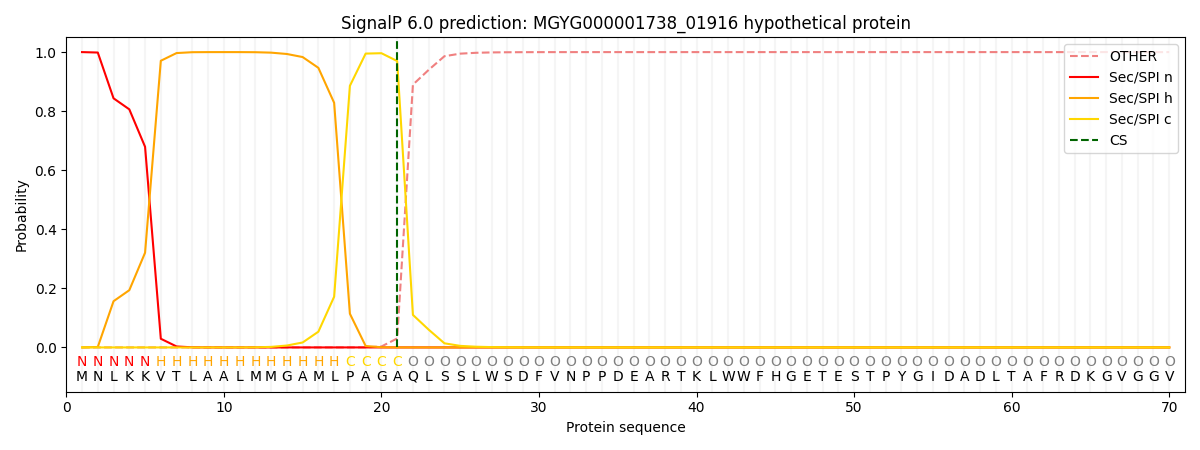
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000388 | 0.998826 | 0.000227 | 0.000216 | 0.000175 | 0.000164 |
