You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001738_02452
You are here: Home > Sequence: MGYG000001738_02452
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Duncaniella sp001701225 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Duncaniella; Duncaniella sp001701225 | |||||||||||
| CAZyme ID | MGYG000001738_02452 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18491; End: 22975 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 1040 | 1311 | 1.5e-38 | 0.8506944444444444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02488 | PLN02488 | 4.71e-11 | 1021 | 1265 | 190 | 417 | probable pectinesterase/pectinesterase inhibitor |
| PLN02313 | PLN02313 | 2.54e-09 | 1041 | 1317 | 291 | 550 | Pectinesterase/pectinesterase inhibitor |
| PLN02432 | PLN02432 | 4.75e-09 | 1148 | 1351 | 101 | 291 | putative pectinesterase |
| COG4677 | PemB | 7.67e-09 | 1041 | 1265 | 100 | 331 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| pfam01095 | Pectinesterase | 2.80e-08 | 1040 | 1342 | 17 | 292 | Pectinesterase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD41145.1 | 0.0 | 3 | 1494 | 10 | 1495 |
| QCD38476.1 | 0.0 | 1 | 1494 | 8 | 1476 |
| QCP72166.1 | 0.0 | 1 | 1494 | 8 | 1476 |
| QNT65238.1 | 0.0 | 381 | 1494 | 15 | 1133 |
| QUT74167.1 | 0.0 | 3 | 1494 | 9 | 1435 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UW0_A | 4.45e-12 | 1023 | 1347 | 26 | 360 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
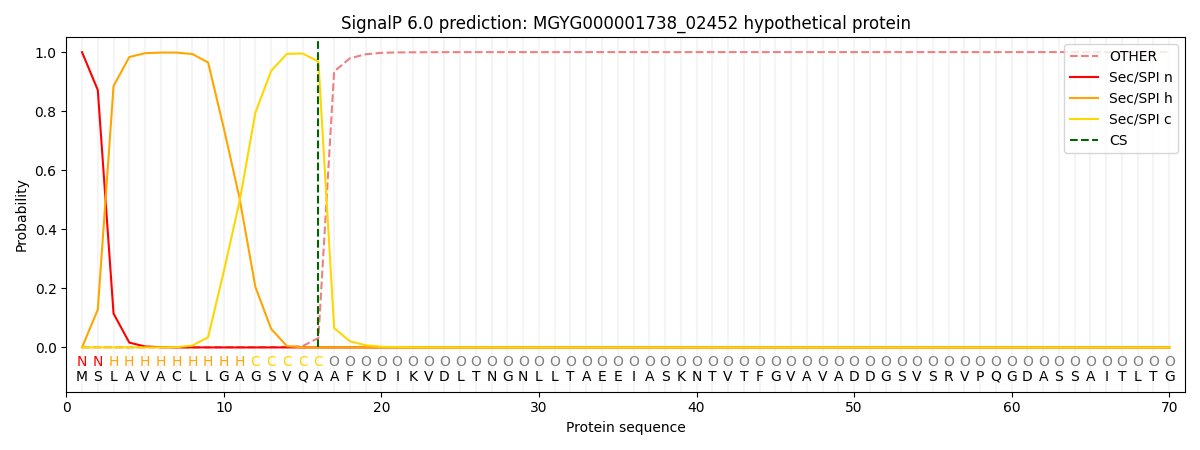
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001157 | 0.997986 | 0.000266 | 0.000208 | 0.000182 | 0.000169 |
