You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001745_01532
You are here: Home > Sequence: MGYG000001745_01532
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
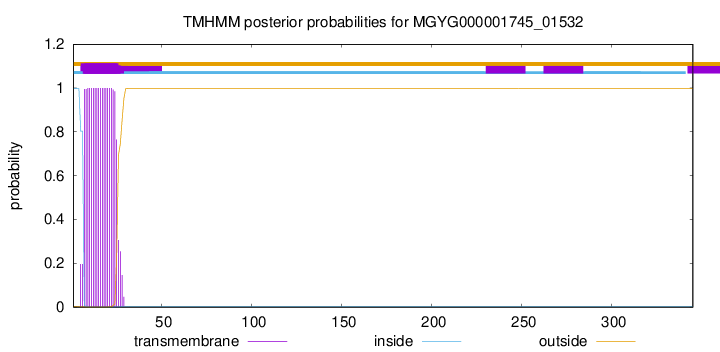
TMHMM annotations
Basic Information help
| Species | Coprococcus sp900548215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Coprococcus; Coprococcus sp900548215 | |||||||||||
| CAZyme ID | MGYG000001745_01532 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 117210; End: 118247 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 62 | 334 | 6.8e-42 | 0.8945454545454545 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.34e-36 | 63 | 321 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.16e-08 | 30 | 253 | 22 | 257 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK83512.1 | 1.18e-188 | 1 | 344 | 1 | 345 |
| QTA37905.1 | 6.09e-89 | 2 | 345 | 3 | 350 |
| AEW47956.1 | 5.66e-69 | 51 | 340 | 55 | 346 |
| AZR73456.1 | 5.29e-54 | 139 | 345 | 1 | 209 |
| QFU77336.1 | 2.92e-33 | 55 | 344 | 73 | 386 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4HTY_A | 2.78e-28 | 58 | 314 | 55 | 323 | CrystalStructure of a metagenome-derived cellulase Cel5A [uncultured bacterium],4HU0_A Crystal Structure of a metagenome-derived cellulase Cel5A in complex with cellotetraose [uncultured bacterium] |
| 3PZT_A | 1.35e-17 | 28 | 334 | 15 | 305 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 5ECU_A | 4.38e-15 | 64 | 323 | 40 | 298 | Theunliganded structure of Caldicellulosiruptor saccharolyticus GH5 [Caldicellulosiruptor saccharolyticus] |
| 1EGZ_A | 3.64e-14 | 125 | 345 | 79 | 285 | ChainA, ENDOGLUCANASE Z [Dickeya chrysanthemi],1EGZ_B Chain B, ENDOGLUCANASE Z [Dickeya chrysanthemi],1EGZ_C Chain C, ENDOGLUCANASE Z [Dickeya chrysanthemi] |
| 5WH8_A | 2.20e-13 | 46 | 323 | 5 | 279 | CellulaseCel5C_n [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22541 | 1.41e-17 | 50 | 289 | 111 | 349 | Endoglucanase A OS=Butyrivibrio fibrisolvens OX=831 GN=celA PE=1 SV=1 |
| P07983 | 1.97e-16 | 54 | 334 | 41 | 310 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P10475 | 2.65e-16 | 54 | 334 | 41 | 310 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P23549 | 1.56e-15 | 54 | 334 | 41 | 310 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
| P07103 | 4.63e-13 | 125 | 345 | 122 | 328 | Endoglucanase Z OS=Dickeya dadantii (strain 3937) OX=198628 GN=celZ PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
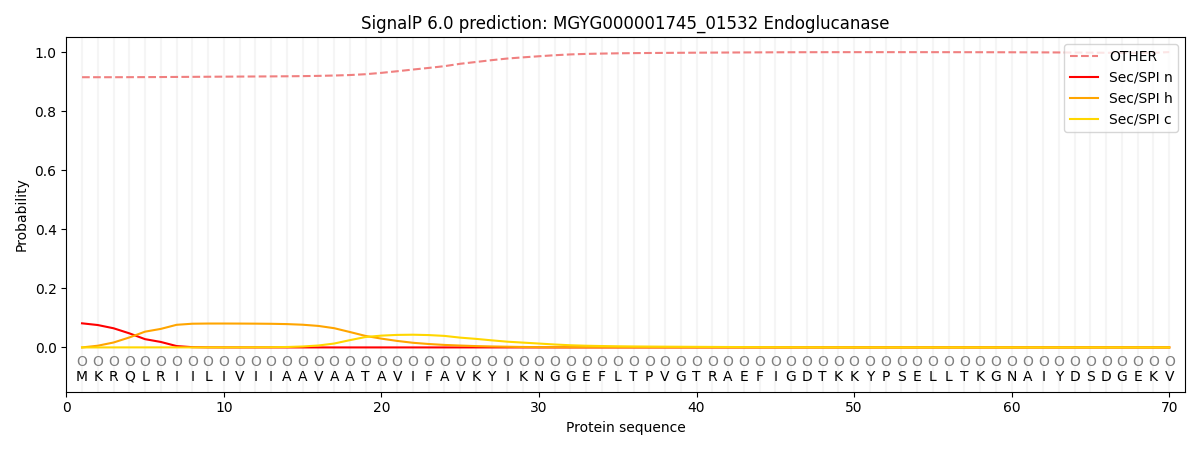
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.919050 | 0.075476 | 0.001991 | 0.000492 | 0.000274 | 0.002747 |