You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001748_00196
You are here: Home > Sequence: MGYG000001748_00196
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
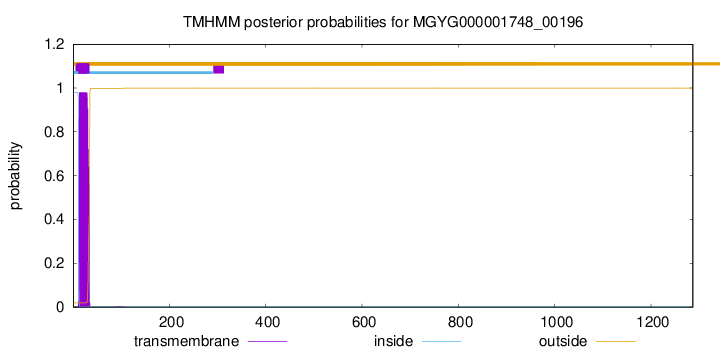
TMHMM annotations
Basic Information help
| Species | CAG-56 sp900762665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-56; CAG-56 sp900762665 | |||||||||||
| CAZyme ID | MGYG000001748_00196 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20832; End: 24695 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 507 | 1003 | 3.2e-64 | 0.5400485436893204 |
| GH106 | 48 | 468 | 8.2e-62 | 0.39805825242718446 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 2.00e-46 | 168 | 799 | 282 | 870 | alpha-L-rhamnosidase. |
| COG5492 | YjdB | 5.78e-22 | 1036 | 1193 | 174 | 324 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam02368 | Big_2 | 1.26e-14 | 1045 | 1118 | 2 | 75 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| smart00635 | BID_2 | 3.99e-10 | 1043 | 1118 | 2 | 80 | Bacterial Ig-like domain 2. |
| COG5492 | YjdB | 3.96e-09 | 1075 | 1196 | 130 | 254 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AND79497.1 | 2.92e-143 | 37 | 1022 | 26 | 995 |
| QTU60664.1 | 1.24e-139 | 44 | 1029 | 40 | 965 |
| QTU52635.1 | 1.24e-139 | 44 | 1029 | 40 | 965 |
| QTU44519.1 | 1.24e-139 | 44 | 1029 | 40 | 965 |
| CBG74689.1 | 1.24e-139 | 44 | 1029 | 40 | 965 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 7.09e-22 | 208 | 1002 | 408 | 1100 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 1.40e-18 | 192 | 1002 | 346 | 1059 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 3.16e-18 | 192 | 1002 | 346 | 1059 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P33747 | 3.75e-13 | 1041 | 1193 | 35 | 190 | Uncharacterized protein CA_P0160 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_P0160 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
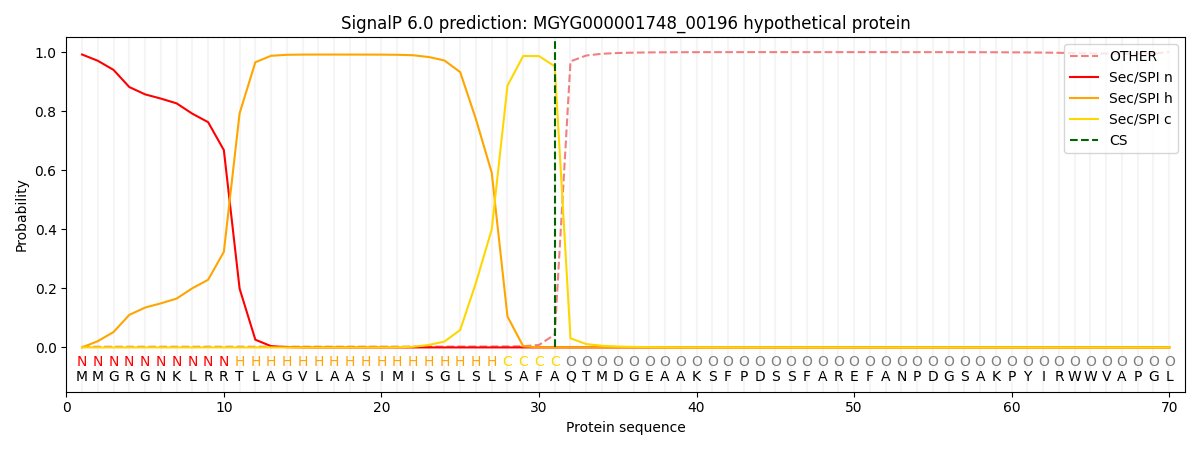
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.005611 | 0.985087 | 0.001579 | 0.006563 | 0.000636 | 0.000459 |