You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001748_00480
You are here: Home > Sequence: MGYG000001748_00480
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-56 sp900762665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-56; CAG-56 sp900762665 | |||||||||||
| CAZyme ID | MGYG000001748_00480 | |||||||||||
| CAZy Family | GH127 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 183147; End: 191294 Strand: + | |||||||||||
Full Sequence Download help
| MKKRIISFLL SITMIAGACF PSMPPVHAAA STQQVQQVQE TGNLNYFSNQ GKLNENAFVQ | 60 |
| LPMGAVEAKD WLKQQLYLQK NGLTGAIHDQ YSLYGPDNGW RGGKGDGWEK GAYYLRGLTS | 120 |
| LAWVLGDEEL KGKAMEWIDF ILDSQRENGF MGPVNDGDGS SDNWDWWPRM VILQVIRDYY | 180 |
| EATEQEGKPD ERVLPFFEKY FRYQLQRLPG KPLNSWAASR GGDNIEVLLW YYNRVYDESD | 240 |
| PTATDWIIDL ASVLASQTKS QDSGLDWNAV FNDTTVREHV VNTTQAMKTP AVLSQLPGHE | 300 |
| GDVNSLKQGI FNMGLDHGRV DNLANADEGA RENYPYRGAE LCSVVESLLS NEMSIRITGE | 360 |
| SWLGDQIEQT AYNNLPAGYA PDYTGHNYFQ AQNQVLGTHG NHEFDCDHGD DSAYGALTGF | 420 |
| ECCFPNMHMG WPKFVQSMWM ATKDNGLAVV AYGPNQVTAK VADGKTAVFE EVTDYPFKDT | 480 |
| IALNYSGDTA KFPLKLRLPA WCEKPEVTIN GVTADLSVQE KGFAVLNREW KPGDQVSVTF | 540 |
| PMKVRTSTWF NNSQAVEYGP LIFAVKVEED WRIGTDDAAK EIQYDPVGEF DRKEVYPASD | 600 |
| WNYGLVLNEA DPEGSFEITA EDEISLQPFI LDNAPITMKV KGQLIPQWKL KGNVVPEPPY | 660 |
| SPIAPDESLQ REIELVPYGC TRIRITQIPV IGEPCSDGVT ERTLEDAQIY EENGTRVTEF | 720 |
| DNIVMPFAED YTLRISYEGS GSMEMNINQK YSESVRFDGS GTMTAENLCE IVPGTNRYFH | 780 |
| FGYGKYNNIR FFGKDVTITK LEILPVDLFT QPEIYSAVIS KDGTSVTLNT NIDRADGFYT | 840 |
| VHYGTESGNY TRTAENFFDK KAVLTGLTPG EDYYFQISML VNGVEKVTEE VKAVAAKEQP | 900 |
| LSFKDDFSDP AASKKQWTLY DPKNVVTFEE GKMSVGSSDN IKAMTGEQQW KDYAVVARLT | 960 |
| GTGKAERDFG IILRASDIGD GSDGYKGYYV GINAIAEGLN IGYADGGWNG IAAPGGITYE | 1020 |
| EGKTYELKTI IAGERLAVYV DGVKLYDELI SNMRSGDRPV PLYESGSAGV RSWNQSFDVN | 1080 |
| SFEVREISQE EYAELGIENH LFEDDFSSAE NSRTKWTVYD PDGAVEFTDG GLKVGNSKNL | 1140 |
| KIMAGTGEEE WENYAVETKL KGPQNPQRDF GVMFRCTDVT EEGADSYFGY YVGIDAIGGG | 1200 |
| LNVGYANGGW NDIEKVRAFQ YEPGKVYDLK ILVCGNQFKV YVDGQQYYEF TDDKFPYGSV | 1260 |
| GLRSWNQPFE AGYFKVRRLT AEEEGQFDQK TDPNVPEEVV PQFSDDFEDT AASAEKWRLC | 1320 |
| GDKSRMKFEA GKLSMGSSDN VKAVAGDENW KDYVAEVSVA LDAKGEQNAG LMYRVTGVEE | 1380 |
| NGSDGYNGYY YGIGNNRDGT GYFIIGYADG DWHQTMRKNL PAFEAGTEYV LKAVVYGDRI | 1440 |
| ALYLDDQMIT RFVDARYASG MIGLRSYDKA FTADNVTVRS LTDADKAGFD GMSRYIEETF | 1500 |
| GAYKTIQLKF PKFSSSKNYK IIYGTEPGNY TNEVYNLNHW KSGSRSDKQG LTLPENDQAY | 1560 |
| YLKLIALDGK EETTVSDEVM VHTGDKPDLS EELEKLSVVL KDAEQTSQDG MTEESSERLN | 1620 |
| WAIAYAKEVA GAPDRNLMEV RLAKNSLLVG KNELEEDTNA EVLPVKVKLE AENAQLAGVA | 1680 |
| KLVERGDASG GYKVGMIDNQ DATVTYTLKA PKDGSYRIEL ATGSGADQPN ASHMYYVNGQ | 1740 |
| KDQAKIVTYQ PNGWDNWSLY PVEVELKKGE NTFTVTHSGR ENSFSELDYI IFYTSNPKVD | 1800 |
| KITLDGEALE GFERDTLTYQ IKVQDLEKLP KVAAELSPDA AEEFEVTVIQ GREASVTLTH | 1860 |
| KKDKTFTLTY QVRFYDDRAF DSSIVNFGAD PYVTYQDGYY YYCRVLQDKA IYVSKAKELN | 1920 |
| RIAATEPQLV YTPAAGEPNR ELWAPEMHYM NGKWYIYYTA GAGADHRMYV LESKTGDALG | 1980 |
| EYEFKGKLAP ETDRWAIDQT VLEYGGKLYA VWSGWEGTTN VSQRIYIAEM SDPLTITGER | 2040 |
| AELSRPEYSW ELDGTPTINE GAQIAVSPEG VVNIIYSASG SWTDNYCLGR LTLRGENPMN | 2100 |
| PEDWEKGTES VFHKSAPSTY STGHACFTKS PDGTEDYIVY HATRGAGQGW NGRGVRTQMF | 2160 |
| TWNEDGTPHF GTALAYDGKV NMPSGTETAK RSRYEAEEGT LLGGAAVEET YNSSGKKKVT | 2220 |
| GLRSDGDGVS FTVEAQTAGI YHLYLGAATK EEGAGLELQV NGAQSVKKNV LPFNAASGGG | 2280 |
| LCPDNWIGYE FSVTLAKGEN NIDVRKMDGQ PMPDLDYLDM ELEEEIKADK TELQEVYDLH | 2340 |
| KGKQETDYTP KSWKAFGEAL DQAKTLLEDE HADQSQVDLA VKALVEAARK LVRKDGQVTE | 2400 |
| EDLQEAVKKA EEAREAAEEA KKLAEQAQAE AKKQKEEAEA AKKAALEAQK RAEEMADASK | 2460 |
| EEREKAQAEA EEARKQAEAA EKLVKEAEKA VQAAEKARKA AEEAAEVSSR EAEAVRAELT | 2520 |
| AAKEEIEKAR EEAQKAREEA DRLRRAAEAS AKEAEEALKA LQEKQKEQET VNSVKVGEIY | 2580 |
| PSGNLKYLVT SVSEKTVSVT GASRKNLKAI TVPASITIEG DTYRVTAISK NAFFRYKRLQ | 2640 |
| KITIGKNVTS IGSKAFYRDA KLKTIVVRAE KLVKIGKNAW KGIYKKAVIR TPQKKKKAYA | 2700 |
| KLLKKSGIAK TVKIK | 2715 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH43 | 1883 | 2168 | 8.5e-99 | 0.9863945578231292 |
| GH127 | 339 | 563 | 3.2e-30 | 0.42366412213740456 |
| CBM66 | 1114 | 1267 | 1.1e-21 | 0.9290322580645162 |
| CBM66 | 929 | 1077 | 1.5e-17 | 0.8580645161290322 |
| CBM35 | 1670 | 1790 | 3.6e-17 | 0.9663865546218487 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd18820 | GH43_LbAraf43-like | 8.95e-127 | 1889 | 2143 | 1 | 258 | Glycosyl hydrolase family 43 proteins similar to Lactobacillus brevis alpha-L-arabinofuranosidase LbAraf43 and Geobacillus thermoleovorans GbtXyl43B. This uncharacterized glycosyl hydrolase family 43 (GH43) subgroup belongs to a subgroup which includes enzymes with beta-xylosidase (EC 3.2.1.37), alpha-L-arabinofuranosidase (EC 3.2.1.55) and possibly bifunctional xylosidase/arabinofuranosidase activities, similar to Lactobacillus brevis alpha-L-arabinofuranosidase LbAraf43 and Geobacillus thermoleovorans IT-08 beta-xylosidase / exo-xylanase (GbtXyl43B). It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd08980 | GH43_LbAraf43-like | 3.05e-76 | 1889 | 2142 | 1 | 275 | Glycosyl hydrolase family 43 proteins such as Lactobacillus brevis alpha-L-arabinofuranosidase LbAraf43 and Geobacillus thermoleovorans GbtXyl43B. This glycosyl hydrolase family 43 (GH43) subgroup includes enzymes with beta-xylosidase (EC 3.2.1.37), alpha-L-arabinofuranosidase (EC 3.2.1.55) and possibly bifunctional xylosidase/arabinofuranosidase activities. In addition to Lactobacillus brevis alpha-L-arabinofuranosidase LbAraf43 and Geobacillus thermoleovorans IT-08 beta-xylosidase / exo-xylanase (GbtXyl43B). It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) familiesGH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd18817 | GH43f_LbAraf43-like | 6.02e-63 | 1889 | 2142 | 1 | 261 | Glycosyl hydrolase family 43 such as Lactobacillus brevis alpha-L-arabinofuranosidase LbAraf43. This glycosyl hydrolase family 43 (GH43) subgroup includes characterized enzymes with alpha-L-arabinofuranosidase (EC 3.2.1.55) activity. It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. Characterized enzymes belonging to this subgroup include Lactobacillus brevis (LbAraf43) and Weissella sp (WAraf43) which show activity with similar catalytic efficiency on 1,5-alpha-L-arabinooligosaccharides with a degree of polymerization (DP) of 2-3; size is limited by an extended loop at the entrance to the active site. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| COG3940 | COG3940 | 1.93e-60 | 1889 | 2175 | 14 | 315 | Beta-xylosidase, GH43 family [Carbohydrate transport and metabolism]. |
| pfam07944 | Glyco_hydro_127 | 6.82e-46 | 106 | 564 | 46 | 503 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJE95537.1 | 1.07e-150 | 49 | 696 | 31 | 653 |
| ALL07583.1 | 2.19e-143 | 39 | 710 | 21 | 676 |
| QPH39723.1 | 1.42e-139 | 19 | 694 | 8 | 652 |
| QNF30112.1 | 4.25e-138 | 48 | 710 | 30 | 676 |
| AMP98056.1 | 4.41e-138 | 46 | 710 | 28 | 676 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M8B_A | 7.56e-44 | 1889 | 2179 | 36 | 346 | ChainA, Alpha-L-arabinofuranosidase II [Levilactobacillus brevis],5M8B_B Chain B, Alpha-L-arabinofuranosidase II [Levilactobacillus brevis] |
| 3K1U_A | 1.06e-42 | 1889 | 2183 | 18 | 327 | Beta-xylosidase,family 43 glycosyl hydrolase from Clostridium acetobutylicum [Clostridium acetobutylicum] |
| 3AKF_A | 5.47e-42 | 1879 | 2175 | 10 | 317 | Crystalstructure of exo-1,5-alpha-L-arabinofuranosidase [Streptomyces avermitilis MA-4680 = NBRC 14893],3AKG_A Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranobiose [Streptomyces avermitilis MA-4680 = NBRC 14893],3AKH_A Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranotriose [Streptomyces avermitilis MA-4680 = NBRC 14893],3AKI_A Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-L-arabinofuranosyl azido [Streptomyces avermitilis MA-4680 = NBRC 14893] |
| 5M8E_A | 3.82e-35 | 1880 | 2171 | 25 | 336 | Crystalstructure of a GH43 arabonofuranosidase from Weissella sp. strain 142 [Weissella cibaria],5M8E_B Crystal structure of a GH43 arabonofuranosidase from Weissella sp. strain 142 [Weissella cibaria] |
| 4QJY_A | 3.21e-14 | 339 | 566 | 332 | 562 | Crystalstructure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QJY_B Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q82P90 | 3.86e-41 | 1879 | 2175 | 36 | 343 | Extracellular exo-alpha-(1->5)-L-arabinofuranosidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=Araf43A PE=1 SV=1 |
| P82594 | 5.78e-35 | 1880 | 2142 | 50 | 318 | Extracellular exo-alpha-(1->5)-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| G4MMH2 | 2.00e-31 | 1879 | 2248 | 40 | 415 | Alpha-L-arabinofuranosidase B OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=abfB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
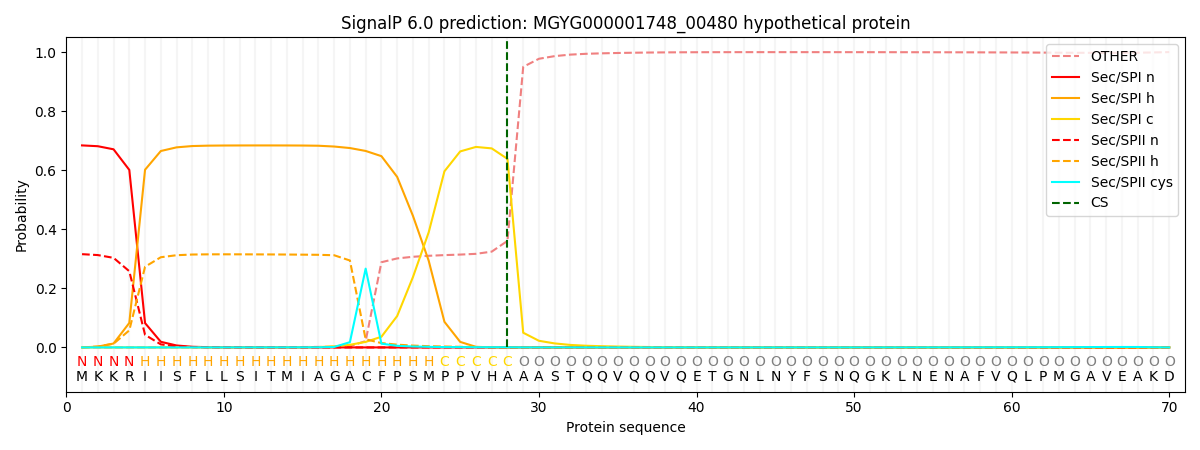
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000614 | 0.673987 | 0.324340 | 0.000480 | 0.000309 | 0.000225 |
