You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001748_00787
You are here: Home > Sequence: MGYG000001748_00787
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-56 sp900762665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-56; CAG-56 sp900762665 | |||||||||||
| CAZyme ID | MGYG000001748_00787 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 145323; End: 149240 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 47 | 1036 | 6.9e-53 | 0.9101941747572816 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PTZ00121 | PTZ00121 | 1.22e-05 | 1151 | 1236 | 1302 | 1385 | MAEBL; Provisional |
| PTZ00121 | PTZ00121 | 2.66e-05 | 1160 | 1262 | 1387 | 1489 | MAEBL; Provisional |
| COG0711 | AtpF | 3.91e-05 | 1163 | 1258 | 50 | 142 | FoF1-type ATP synthase, membrane subunit b or b' [Energy production and conversion]. |
| NF033777 | M_group_A_cterm | 8.78e-05 | 1162 | 1251 | 24 | 131 | M protein C-terminal domain. M protein (emm) is an important virulence protein and serology-defining surface antigen of Streptococcus pyogenes (group A Streptococcus). M protein has an amino-terminal YSIRK-type signal sequence (associated with cross-wall targeting in dividing cells), and a C-terminal LPXTG domain for processing by sortase and covalent attachment to the Gram-positive cell wall. Past the signal peptide, M protein has a hypervariable region, but this HMM describes only the well-conserved region C-terminal to the hypervariable region. It discriminates M protein from two related proteins, Enn and Mrp. |
| pfam07321 | YscO | 1.53e-04 | 1164 | 1210 | 76 | 119 | Type III secretion protein YscO. This family contains the bacterial type III secretion protein YscO, which is approximately 150 residues long. YscO has been shown to be required for high-level expression and secretion of the anti-host proteins V antigen and Yops in Yersinia pestis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AHF25191.1 | 0.0 | 42 | 1155 | 57 | 1152 |
| QGH71057.1 | 0.0 | 13 | 1152 | 48 | 1128 |
| QGH70002.1 | 4.31e-103 | 34 | 1037 | 73 | 1054 |
| QYN20448.1 | 7.83e-101 | 52 | 1111 | 57 | 1087 |
| SDS67158.1 | 9.91e-80 | 52 | 1041 | 12 | 912 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
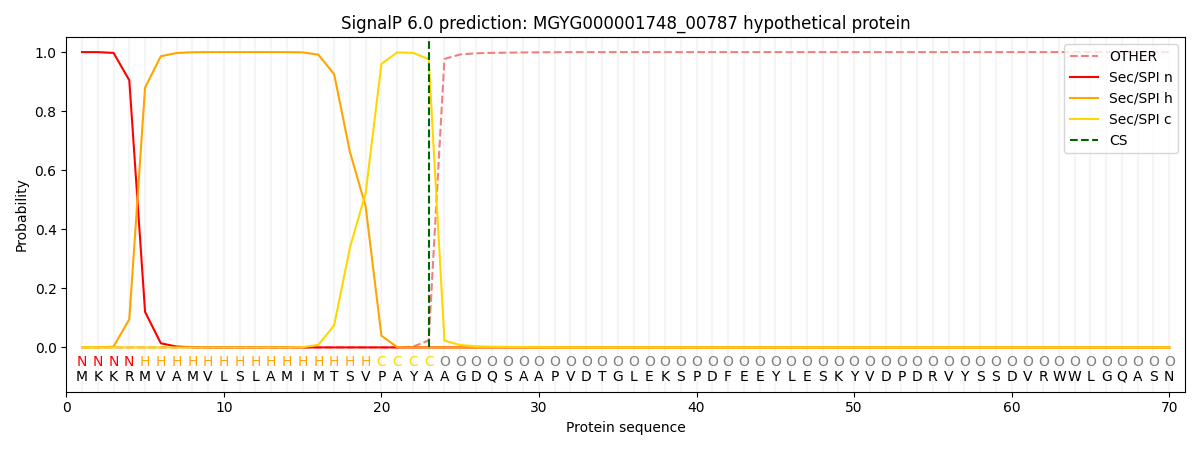
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000270 | 0.998985 | 0.000186 | 0.000208 | 0.000183 | 0.000158 |
