You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001750_01248
You are here: Home > Sequence: MGYG000001750_01248
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola ilei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola ilei | |||||||||||
| CAZyme ID | MGYG000001750_01248 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29545; End: 32409 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 21 | 721 | 1.7e-82 | 0.7021276595744681 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.27e-29 | 17 | 479 | 6 | 433 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 3.07e-28 | 26 | 455 | 15 | 421 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.60e-17 | 117 | 451 | 113 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02837 | Glyco_hydro_2_N | 2.78e-17 | 26 | 218 | 4 | 169 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam00703 | Glyco_hydro_2 | 2.73e-10 | 221 | 325 | 2 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AAO79256.1 | 0.0 | 26 | 954 | 27 | 957 |
| QMW86500.1 | 0.0 | 26 | 954 | 27 | 957 |
| QUT42008.1 | 0.0 | 26 | 954 | 27 | 957 |
| BCA48689.1 | 0.0 | 26 | 954 | 27 | 957 |
| ALJ43488.1 | 0.0 | 26 | 954 | 27 | 957 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3BGA_A | 7.10e-25 | 26 | 474 | 51 | 485 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 3DEC_A | 4.81e-24 | 75 | 474 | 85 | 481 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 6XXW_A | 5.27e-21 | 111 | 453 | 70 | 418 | Structureof beta-D-Glucuronidase for Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
| 6D8K_A | 3.73e-20 | 106 | 479 | 90 | 443 | Bacteroidesmultiple species beta-glucuronidase [Bacteroides ovatus],6D8K_B Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_C Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_D Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus] |
| 3K4A_A | 6.24e-19 | 26 | 452 | 17 | 416 | Crystalstructure of selenomethionine substituted E. coli beta-glucuronidase [Escherichia coli K-12],3K4A_B Crystal structure of selenomethionine substituted E. coli beta-glucuronidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KPJ7 | 4.30e-19 | 105 | 463 | 96 | 437 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| P77989 | 3.49e-18 | 98 | 445 | 41 | 381 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| P05804 | 4.49e-18 | 26 | 452 | 15 | 414 | Beta-glucuronidase OS=Escherichia coli (strain K12) OX=83333 GN=uidA PE=1 SV=2 |
| O52847 | 2.11e-16 | 17 | 454 | 55 | 484 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| P24131 | 1.72e-15 | 26 | 530 | 49 | 525 | Beta-galactosidase OS=Clostridium acetobutylicum OX=1488 GN=cbgA PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
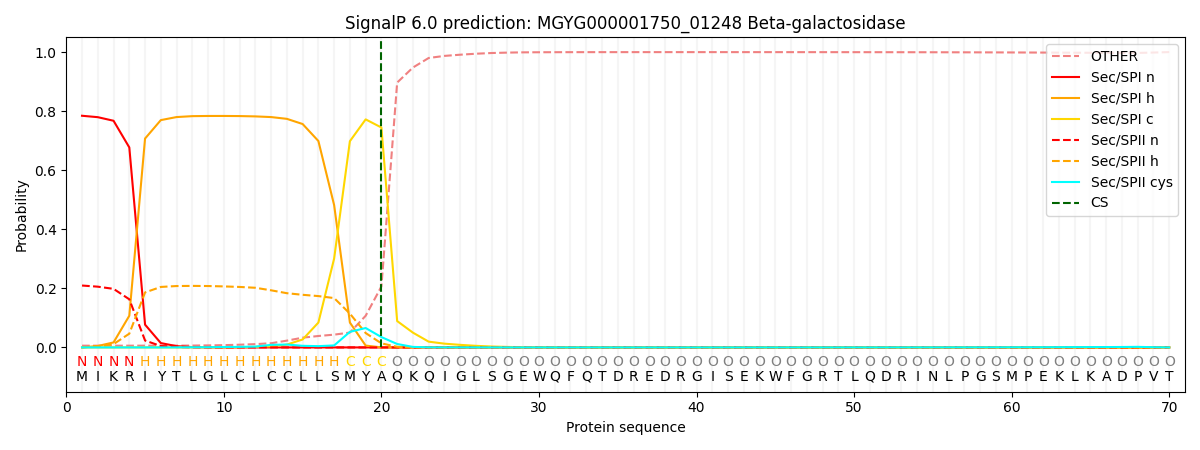
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.009368 | 0.769935 | 0.219622 | 0.000327 | 0.000336 | 0.000374 |
