You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001750_01429
You are here: Home > Sequence: MGYG000001750_01429
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola ilei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola ilei | |||||||||||
| CAZyme ID | MGYG000001750_01429 | |||||||||||
| CAZy Family | GH143 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29610; End: 32954 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH143 | 29 | 577 | 1.3e-282 | 0.987410071942446 |
| GH142 | 640 | 1106 | 1.1e-219 | 0.9958246346555324 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13088 | BNR_2 | 1.24e-08 | 272 | 346 | 168 | 247 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| COG3408 | GDB1 | 0.003 | 764 | 968 | 346 | 551 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| cd08995 | GH32_EcAec43-like | 0.007 | 83 | 130 | 3 | 52 | Glycosyl hydrolase family 32, such as the putative glycoside hydrolase Escherichia coli Aec43 (FosGH2). This glycosyl hydrolase family 32 (GH32) subgroup includes Escherichia coli strain BEN2908 putative glycoside hydrolase Aec43 (FosGH2). GH32 enzymes cleave sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). GH32 family also contains other fructofuranosidases such as inulinase (EC 3.2.1.7), exo-inulinase (EC 3.2.1.80), levanase (EC 3.2.1.65), and transfructosidases such sucrose:sucrose 1-fructosyltransferase (EC 2.4.1.99), fructan:fructan 1-fructosyltransferase (EC 2.4.1.100), sucrose:fructan 6-fructosyltransferase (EC 2.4.1.10), fructan:fructan 6G-fructosyltransferase (EC 2.4.1.243) and levan fructosyltransferases (EC 2.4.1.-). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. These enzymes are predicted to display a 5-fold beta-propeller fold as found for GH43 and CH68. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJR63716.1 | 0.0 | 26 | 1114 | 18 | 1103 |
| QJR67979.1 | 0.0 | 26 | 1114 | 18 | 1103 |
| QJR72315.1 | 0.0 | 26 | 1114 | 18 | 1103 |
| QJR59518.1 | 0.0 | 26 | 1114 | 18 | 1103 |
| QJR76384.1 | 0.0 | 26 | 1114 | 18 | 1103 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQR_A | 0.0 | 31 | 1114 | 26 | 1107 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 5MQS_A | 0.0 | 31 | 1114 | 26 | 1107 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 6M5A_A | 8.62e-23 | 640 | 1109 | 284 | 810 | Crystalstructure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 7.63e-22 | 640 | 1109 | 315 | 841 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
| P94250 | 1.80e-08 | 764 | 945 | 119 | 298 | Uncharacterized protein BB_0381 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0381 PE=4 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
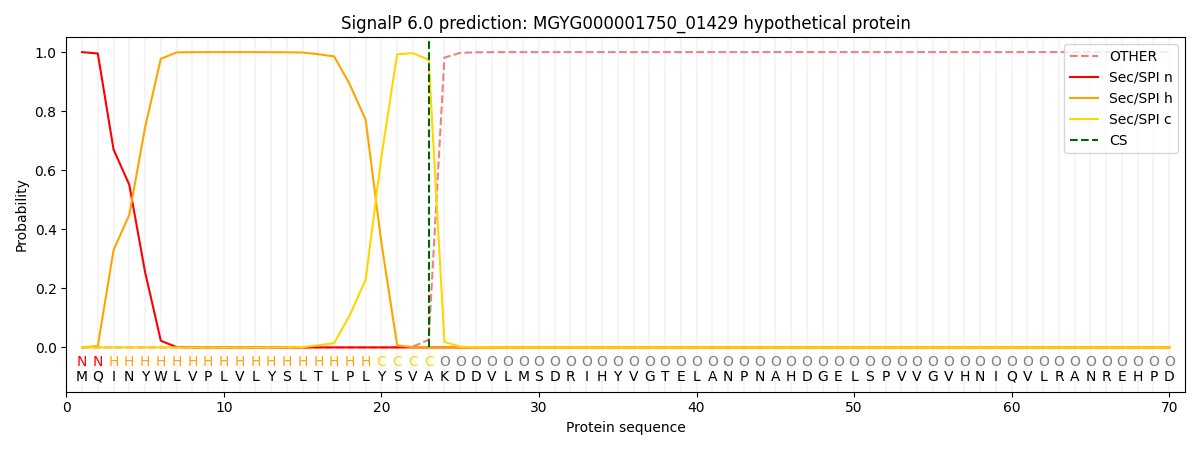
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000294 | 0.999047 | 0.000189 | 0.000139 | 0.000141 | 0.000142 |
