You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001756_01402
You are here: Home > Sequence: MGYG000001756_01402
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
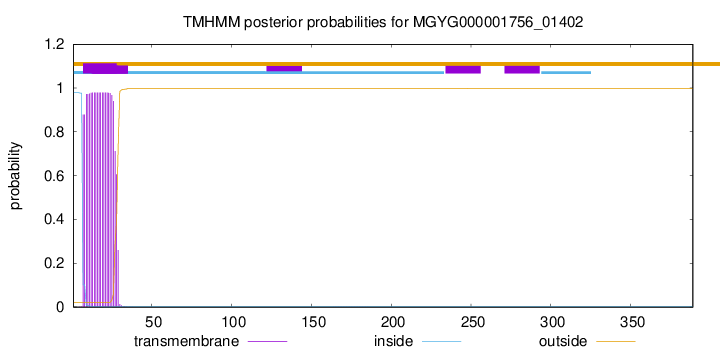
TMHMM annotations
Basic Information help
| Species | UBA1394 sp900538575 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UBA1394; UBA1394 sp900538575 | |||||||||||
| CAZyme ID | MGYG000001756_01402 | |||||||||||
| CAZy Family | CE3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 56511; End: 57680 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 34 | 252 | 8.5e-49 | 0.9948453608247423 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01833 | XynB_like | 2.73e-42 | 34 | 252 | 1 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| pfam13472 | Lipase_GDSL_2 | 2.78e-18 | 39 | 243 | 2 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| pfam00657 | Lipase_GDSL | 2.91e-13 | 36 | 248 | 1 | 224 | GDSL-like Lipase/Acylhydrolase. |
| cd14256 | Dockerin_I | 3.81e-13 | 331 | 387 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| cd00229 | SGNH_hydrolase | 1.13e-12 | 39 | 250 | 4 | 186 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEV68498.1 | 5.32e-41 | 31 | 252 | 135 | 326 |
| CAB55348.1 | 6.39e-38 | 29 | 267 | 39 | 279 |
| AEY65113.1 | 1.99e-36 | 36 | 256 | 119 | 309 |
| AVT35642.1 | 6.12e-35 | 28 | 252 | 30 | 227 |
| BCJ76402.1 | 6.61e-35 | 25 | 267 | 28 | 243 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VPT_A | 9.35e-21 | 34 | 249 | 6 | 192 | ChainA, LIPOLYTIC ENZYME [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9RLB8 | 1.28e-38 | 29 | 267 | 39 | 279 | Multidomain esterase OS=Ruminococcus flavefaciens OX=1265 GN=cesA PE=1 SV=1 |
| P15329 | 1.16e-08 | 94 | 264 | 1 | 151 | Putative endoglucanase X (Fragment) OS=Acetivibrio thermocellus OX=1515 GN=celX PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000272 | 0.999099 | 0.000179 | 0.000163 | 0.000138 | 0.000126 |