You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001765_00747
You are here: Home > Sequence: MGYG000001765_00747
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
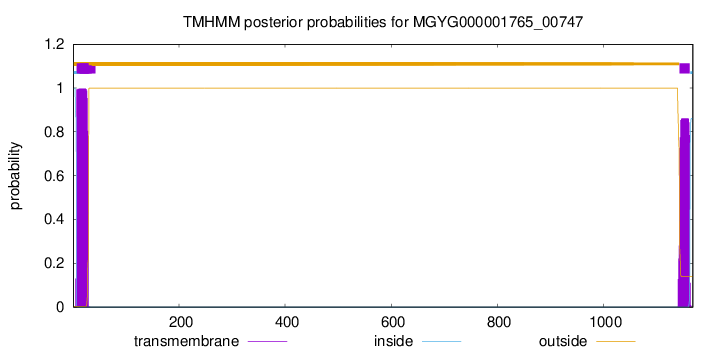
TMHMM annotations
Basic Information help
| Species | CAG-448 sp000433415 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; CAG-272; CAG-448; CAG-448 sp000433415 | |||||||||||
| CAZyme ID | MGYG000001765_00747 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 27252; End: 30761 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 701 | 830 | 4.7e-16 | 0.9354838709677419 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 6.74e-12 | 70 | 183 | 9 | 122 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 4.26e-10 | 888 | 988 | 11 | 114 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 9.89e-08 | 701 | 822 | 2 | 117 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 0.002 | 700 | 833 | 13 | 142 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SDS28274.1 | 2.90e-134 | 37 | 682 | 61 | 693 |
| QBR87893.1 | 1.15e-107 | 60 | 681 | 221 | 834 |
| QOR71214.1 | 3.19e-104 | 30 | 689 | 204 | 851 |
| QNK60264.1 | 5.11e-74 | 62 | 836 | 47 | 878 |
| SDS02659.1 | 1.90e-69 | 73 | 835 | 224 | 1020 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
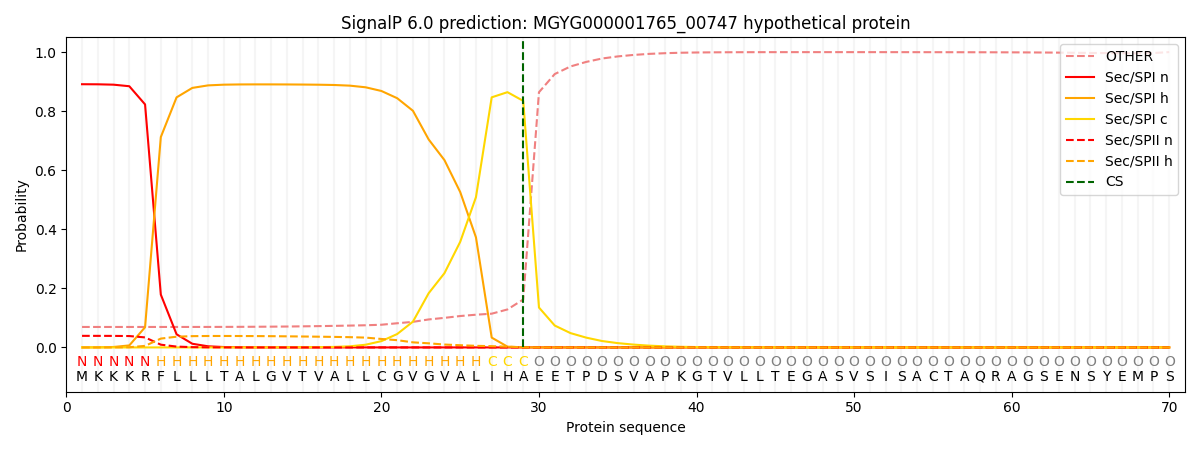
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.076587 | 0.878526 | 0.041937 | 0.001916 | 0.000565 | 0.000437 |