You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001767_00226
You are here: Home > Sequence: MGYG000001767_00226
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp000432655 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp000432655 | |||||||||||
| CAZyme ID | MGYG000001767_00226 | |||||||||||
| CAZy Family | GH54 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8709; End: 11327 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH54 | 34 | 351 | 5.2e-119 | 0.990506329113924 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam09206 | ArabFuran-catal | 7.07e-145 | 34 | 350 | 1 | 314 | Alpha-L-arabinofuranosidase B, catalytic. Members of this family, which are present in fungal alpha-L-arabinofuranosidase B, adopt a beta-sandwich fold similar to that of Concanavalin A-like lectins/glucanase. The beta-sandwich fold consists of two anti-parallel beta-sheets with seven and and six strands, respectively. In addition, there are four helices outside of the beta-strands. The beta-sandwich strands are closely packed and curved with a jelly roll topology, creating a small catalytic pocket. The domain catalyzes the hydrolysis of alpha-1,2-, alpha-1,3- and alpha-1,5-L-arabinofuranosidic bonds in L-arabinose-containing hemicelluloses such as arabinoxylan and L-arabinan. |
| pfam00932 | LTD | 1.14e-09 | 450 | 525 | 6 | 72 | Lamin Tail Domain. The lamin-tail domain (LTD), which has an immunoglobulin (Ig) fold, is found in Nuclear Lamins, Chlo1887 from Chloroflexus, and several bacterial proteins where it occurs with membrane associated hydrolases of the metallo-beta-lactamase,synaptojanin, and calcineurin-like phosphoesterase superfamilies. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOY90220.1 | 2.82e-237 | 12 | 861 | 6 | 867 |
| AJP74471.1 | 4.37e-194 | 57 | 859 | 3 | 818 |
| ACB75541.1 | 8.69e-143 | 27 | 711 | 39 | 768 |
| QOY90219.1 | 1.01e-139 | 27 | 743 | 34 | 805 |
| AXC15854.1 | 8.90e-139 | 27 | 744 | 41 | 820 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1WD3_A | 1.14e-104 | 34 | 350 | 3 | 316 | Crystalstructure of arabinofuranosidase [Aspergillus luchuensis],1WD4_A Crystal structure of arabinofuranosidase complexed with arabinose [Aspergillus luchuensis],6SXS_AAA Chain AAA, Alpha-L-arabinofuranosidase B [Aspergillus luchuensis] |
| 6SXT_A | 1.17e-104 | 34 | 350 | 4 | 317 | GH54a-l-arabinofuranosidase soaked with aziridine inhibitor [Aspergillus luchuensis IFO 4308] |
| 6SXR_A | 3.13e-104 | 34 | 350 | 3 | 316 | E221Qmutant of GH54 a-l-arabinofuranosidase soaked with 4-nitrophenyl a-l-arabinofuranoside [Aspergillus luchuensis IFO 4308] |
| 2D43_A | 8.61e-104 | 34 | 350 | 3 | 316 | ChainA, alpha-L-arabinofuranosidase B [Aspergillus luchuensis],2D44_A Chain A, alpha-L-arabinofuranosidase B [Aspergillus luchuensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1CGD1 | 1.53e-111 | 2 | 350 | 7 | 341 | Probable alpha-L-arabinofuranosidase B OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=abfB PE=3 SV=1 |
| Q92455 | 4.93e-111 | 34 | 355 | 23 | 340 | Alpha-L-arabinofuranosidase OS=Hypocrea jecorina OX=51453 GN=abf1 PE=2 SV=1 |
| O74288 | 9.38e-111 | 34 | 350 | 26 | 340 | Alpha-L-arabinofuranosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=abfB PE=1 SV=1 |
| B8N7I7 | 3.22e-110 | 34 | 350 | 28 | 341 | Probable alpha-L-arabinofuranosidase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=abfB PE=3 SV=1 |
| P48792 | 1.46e-109 | 34 | 355 | 23 | 340 | Arabinofuranosidase/B-xylosidase OS=Trichoderma koningii OX=97093 GN=xyl1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
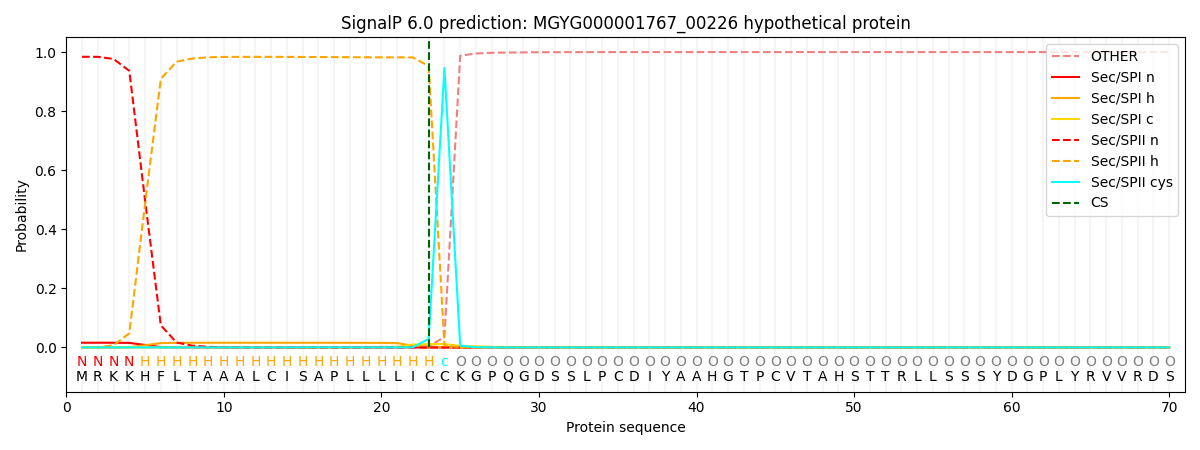
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000021 | 0.015432 | 0.984552 | 0.000005 | 0.000008 | 0.000006 |
