You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001767_00729
You are here: Home > Sequence: MGYG000001767_00729
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp000432655 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp000432655 | |||||||||||
| CAZyme ID | MGYG000001767_00729 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | Reducing end xylose-releasing exo-oligoxylanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34548; End: 35780 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3405 | BcsZ | 1.87e-41 | 23 | 408 | 4 | 350 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| pfam01270 | Glyco_hydro_8 | 2.71e-20 | 74 | 345 | 28 | 284 | Glycosyl hydrolases family 8. |
| PRK11097 | PRK11097 | 2.85e-09 | 70 | 172 | 42 | 146 | cellulase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT88744.1 | 1.22e-157 | 25 | 408 | 34 | 414 |
| ALJ60263.1 | 2.46e-157 | 25 | 408 | 34 | 414 |
| CCH00339.1 | 1.63e-155 | 19 | 408 | 27 | 413 |
| QSB29356.1 | 6.38e-155 | 24 | 408 | 22 | 403 |
| ADB37459.1 | 1.03e-154 | 27 | 408 | 48 | 426 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5YXT_A | 5.84e-84 | 27 | 408 | 13 | 375 | Glycosidehydrolase family 8 Xylanase [Paenibacillus barengoltzii G22],5YXT_B Glycoside hydrolase family 8 Xylanase [Paenibacillus barengoltzii G22],5YXT_C Glycoside hydrolase family 8 Xylanase [Paenibacillus barengoltzii G22],5YXT_D Glycoside hydrolase family 8 Xylanase [Paenibacillus barengoltzii G22] |
| 1WU4_A | 3.81e-79 | 25 | 408 | 12 | 378 | ChainA, xylanase Y [Halalkalibacterium halodurans C-125],1WU5_A Chain A, xylanase Y [Halalkalibacterium halodurans C-125] |
| 3A3V_A | 1.51e-78 | 25 | 408 | 12 | 378 | ChainA, Xylanase Y [Halalkalibacterium halodurans] |
| 2DRR_A | 2.13e-78 | 25 | 408 | 12 | 378 | ChainA, Xylanase Y [Halalkalibacterium halodurans C-125] |
| 1WU6_A | 3.00e-78 | 25 | 408 | 12 | 378 | ChainA, xylanase Y [Halalkalibacterium halodurans C-125] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9KB30 | 1.66e-78 | 25 | 408 | 12 | 378 | Reducing end xylose-releasing exo-oligoxylanase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=BH2105 PE=1 SV=1 |
| A0A0S2UQQ5 | 1.62e-76 | 27 | 408 | 14 | 379 | Reducing-end xylose-releasing exo-oligoxylanase Rex8A OS=Paenibacillus barcinonensis OX=198119 GN=rex8A PE=1 SV=1 |
| A1A048 | 8.79e-76 | 31 | 409 | 16 | 379 | Reducing end xylose-releasing exo-oligoxylanase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=xylA PE=1 SV=1 |
| A3DC29 | 2.22e-23 | 37 | 406 | 51 | 388 | Endoglucanase A OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celA PE=1 SV=1 |
| P37701 | 1.65e-22 | 75 | 406 | 93 | 389 | Endoglucanase 2 OS=Ruminiclostridium josui OX=1499 GN=celB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
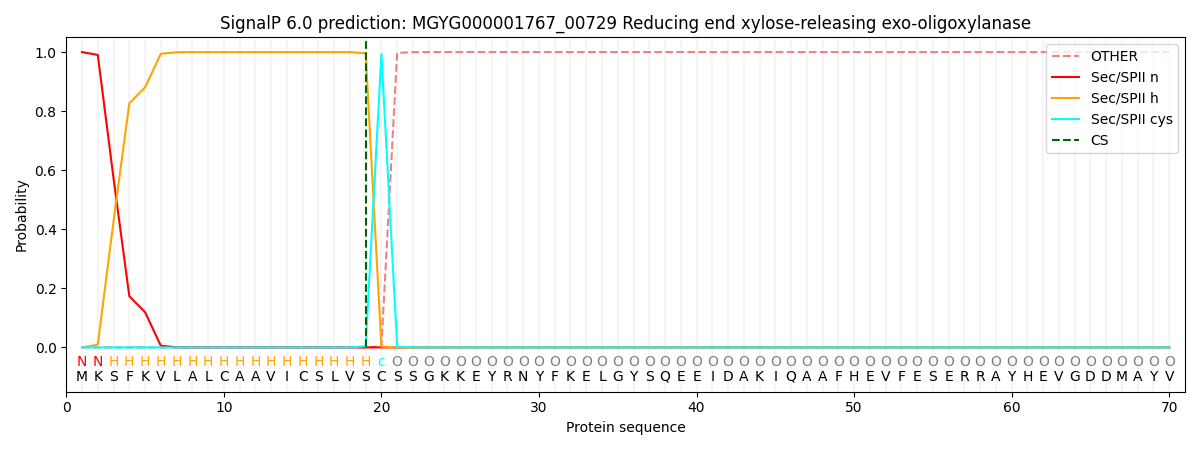
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000002 | 1.000059 | 0.000000 | 0.000000 | 0.000000 |
