You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001770_00889
You are here: Home > Sequence: MGYG000001770_00889
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900313215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900313215 | |||||||||||
| CAZyme ID | MGYG000001770_00889 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | Pectinesterase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8713; End: 9702 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 30 | 324 | 1.1e-100 | 0.9826388888888888 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01095 | Pectinesterase | 1.25e-76 | 31 | 327 | 4 | 298 | Pectinesterase. |
| PLN02773 | PLN02773 | 1.43e-73 | 29 | 317 | 7 | 289 | pectinesterase |
| COG4677 | PemB | 8.16e-67 | 38 | 324 | 93 | 402 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02682 | PLN02682 | 4.28e-63 | 19 | 315 | 61 | 352 | pectinesterase family protein |
| PLN02432 | PLN02432 | 4.98e-60 | 31 | 329 | 15 | 291 | putative pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT66521.1 | 2.75e-194 | 1 | 329 | 1 | 329 |
| AGB28242.1 | 1.10e-162 | 1 | 324 | 1 | 315 |
| BCS84477.1 | 1.12e-160 | 1 | 324 | 1 | 315 |
| QVJ81978.1 | 6.05e-155 | 1 | 329 | 1 | 320 |
| ADE82952.1 | 6.05e-155 | 1 | 329 | 1 | 320 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1XG2_A | 2.78e-39 | 31 | 329 | 7 | 303 | ChainA, Pectinesterase 1 [Solanum lycopersicum] |
| 1GQ8_A | 3.20e-37 | 31 | 306 | 11 | 281 | Pectinmethylesterase from Carrot [Daucus carota] |
| 5C1E_A | 6.52e-33 | 26 | 303 | 8 | 271 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 5C1C_A | 1.27e-32 | 26 | 303 | 8 | 271 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 3UW0_A | 2.28e-30 | 38 | 324 | 43 | 358 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9LVQ0 | 1.16e-58 | 31 | 315 | 9 | 287 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
| P41510 | 2.01e-50 | 28 | 322 | 272 | 565 | Probable pectinesterase/pectinesterase inhibitor OS=Brassica napus OX=3708 GN=BP19 PE=2 SV=1 |
| Q5MFV6 | 8.02e-49 | 28 | 329 | 276 | 576 | Probable pectinesterase/pectinesterase inhibitor VGDH2 OS=Arabidopsis thaliana OX=3702 GN=VGDH2 PE=2 SV=2 |
| Q42608 | 3.25e-48 | 28 | 322 | 259 | 552 | Pectinesterase/pectinesterase inhibitor (Fragment) OS=Brassica campestris OX=3711 PE=2 SV=1 |
| Q8VYZ3 | 6.37e-45 | 19 | 315 | 75 | 366 | Probable pectinesterase 53 OS=Arabidopsis thaliana OX=3702 GN=PME53 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
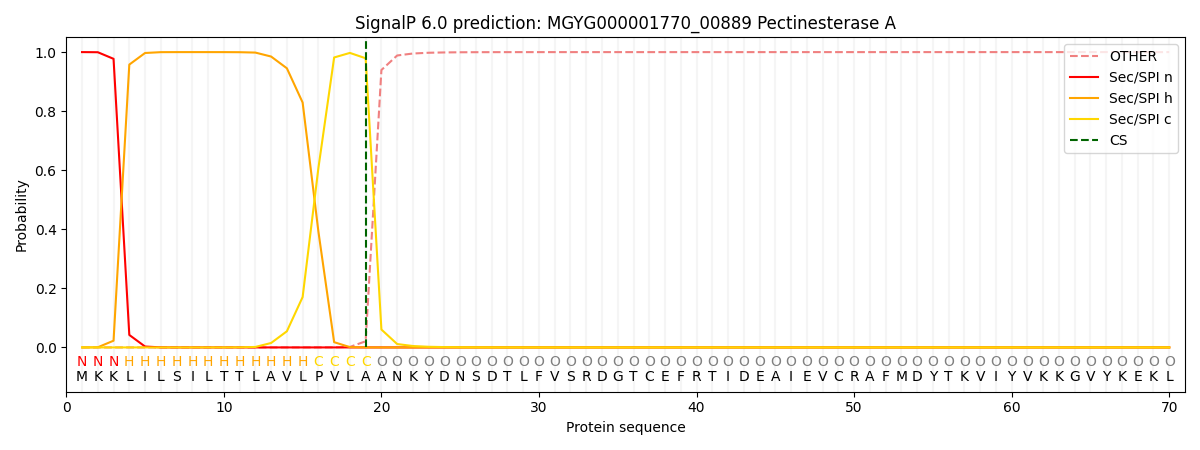
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000288 | 0.999052 | 0.000184 | 0.000162 | 0.000158 | 0.000146 |
