You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001771_00323
You are here: Home > Sequence: MGYG000001771_00323
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-617 sp000438115 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; CAG-617; CAG-617 sp000438115 | |||||||||||
| CAZyme ID | MGYG000001771_00323 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20002; End: 22128 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 22 | 543 | 3.6e-97 | 0.9905123339658444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 2.80e-11 | 358 | 563 | 8 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 1.42e-09 | 331 | 439 | 28 | 120 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 1.32e-06 | 413 | 591 | 2 | 134 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam05048 | NosD | 6.21e-06 | 382 | 575 | 25 | 181 | Periplasmic copper-binding protein (NosD). NosD is a periplasmic protein which is thought to insert copper into the exported reductase apoenzyme (NosZ). This region forms a parallel beta helix domain. |
| pfam07602 | DUF1565 | 3.46e-05 | 28 | 92 | 2 | 63 | Protein of unknown function (DUF1565). These proteins share a region of homology in their N termini, and are found in several phylogenetically diverse bacteria and in the archaeon Methanosarcina acetivorans. Some of these proteins also contain characterized domains such as pfam00395 and pfam03422. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA25764.1 | 4.26e-199 | 1 | 693 | 1 | 677 |
| AKP49975.1 | 1.80e-192 | 21 | 708 | 36 | 713 |
| AEL24383.1 | 7.19e-192 | 22 | 698 | 37 | 703 |
| SCM58138.1 | 8.07e-191 | 14 | 703 | 14 | 702 |
| AWO00314.1 | 3.01e-186 | 18 | 705 | 32 | 700 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQP_A | 1.81e-43 | 23 | 540 | 27 | 580 | Glycosidehydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
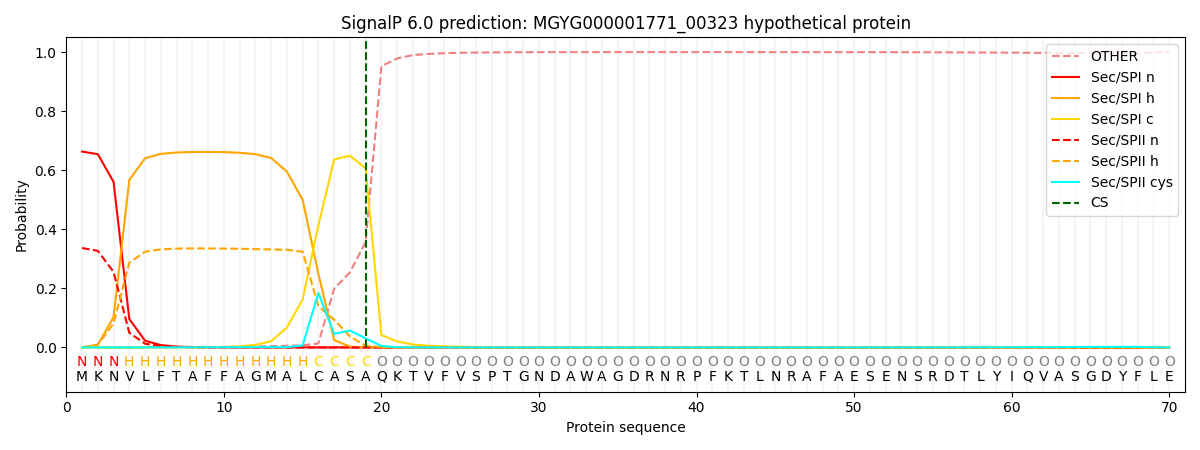
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001297 | 0.650702 | 0.347079 | 0.000356 | 0.000277 | 0.000254 |
