You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001779_01436
You are here: Home > Sequence: MGYG000001779_01436
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
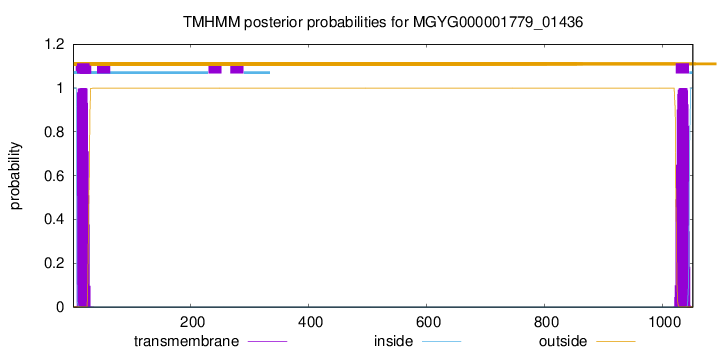
TMHMM annotations
Basic Information help
| Species | UMGS1851 sp900555605 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; UBA1212; UBA1255; UMGS1851; UMGS1851 sp900555605 | |||||||||||
| CAZyme ID | MGYG000001779_01436 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6819; End: 9977 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 355 | 649 | 6e-24 | 0.6861538461538461 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 2.16e-25 | 309 | 601 | 88 | 369 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 9.53e-05 | 411 | 601 | 48 | 210 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23464.1 | 4.19e-178 | 42 | 757 | 71 | 752 |
| QCD41820.1 | 3.39e-109 | 89 | 688 | 248 | 843 |
| QYH35737.1 | 5.83e-44 | 302 | 813 | 126 | 627 |
| ADB48561.1 | 7.07e-40 | 300 | 794 | 26 | 484 |
| QGQ94767.1 | 8.42e-39 | 300 | 804 | 35 | 514 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MXN_A | 2.30e-13 | 307 | 497 | 25 | 190 | Crystalstructure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_B Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_C Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_D Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184] |
| 3JUR_A | 7.58e-11 | 297 | 534 | 23 | 267 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
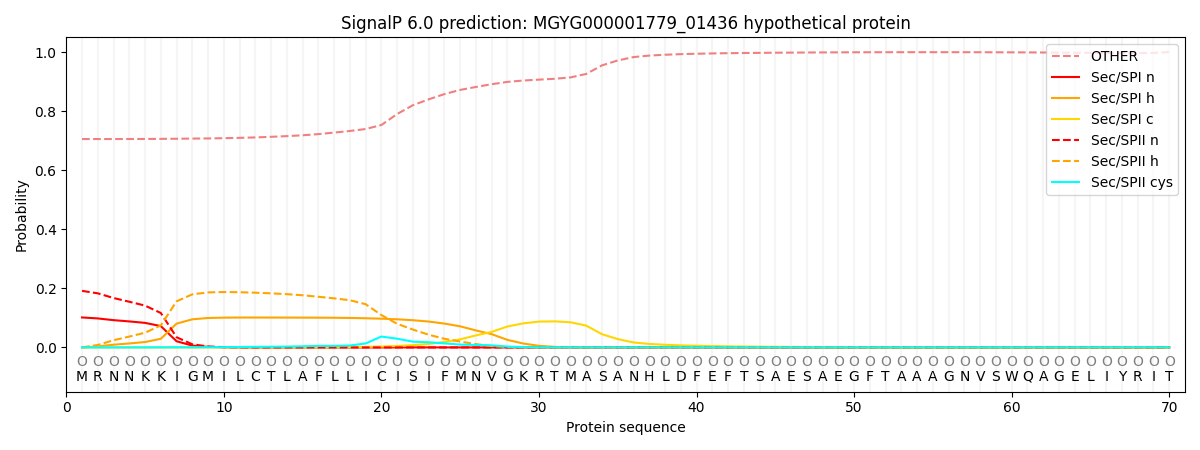
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.709967 | 0.094316 | 0.192572 | 0.000644 | 0.000426 | 0.002091 |