You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001780_03177
You are here: Home > Sequence: MGYG000001780_03177
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; | |||||||||||
| CAZyme ID | MGYG000001780_03177 | |||||||||||
| CAZy Family | GH27 | |||||||||||
| CAZyme Description | Alpha-galactosidase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1416; End: 2636 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH27 | 125 | 378 | 5e-84 | 0.982532751091703 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14792 | GH27 | 1.49e-149 | 30 | 301 | 1 | 271 | glycosyl hydrolase family 27 (GH27). GH27 enzymes occur in eukaryotes, prokaryotes, and archaea with a wide range of hydrolytic activities, including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-N-acetylgalactosaminidase, and 3-alpha-isomalto-dextranase. All GH27 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH27 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| PLN02808 | PLN02808 | 2.04e-130 | 26 | 400 | 28 | 384 | alpha-galactosidase |
| PLN02229 | PLN02229 | 7.11e-129 | 5 | 400 | 31 | 418 | alpha-galactosidase |
| PLN02692 | PLN02692 | 1.30e-120 | 26 | 399 | 52 | 408 | alpha-galactosidase |
| pfam16499 | Melibiase_2 | 4.12e-103 | 29 | 301 | 1 | 284 | Alpha galactosidase A. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFN75841.1 | 6.20e-211 | 2 | 399 | 5 | 404 |
| BBM73909.1 | 4.53e-203 | 19 | 399 | 18 | 398 |
| BBM70930.1 | 4.53e-203 | 19 | 399 | 18 | 398 |
| AEN74501.1 | 1.29e-202 | 19 | 399 | 18 | 398 |
| ACY49470.1 | 1.84e-202 | 19 | 399 | 18 | 398 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1UAS_A | 6.39e-111 | 23 | 402 | 1 | 362 | ChainA, alpha-galactosidase [Oryza sativa] |
| 6F4C_B | 1.44e-104 | 24 | 400 | 3 | 361 | Nicotianabenthamiana alpha-galactosidase [Nicotiana benthamiana] |
| 4OGZ_A | 3.46e-99 | 24 | 339 | 94 | 424 | Crystalstructure of a putative alpha-galactosidase/melibiase (BF4189) from Bacteroides fragilis NCTC 9343 at 2.00 A resolution [Bacteroides fragilis NCTC 9343],4OGZ_B Crystal structure of a putative alpha-galactosidase/melibiase (BF4189) from Bacteroides fragilis NCTC 9343 at 2.00 A resolution [Bacteroides fragilis NCTC 9343] |
| 3A5V_A | 3.56e-97 | 24 | 398 | 3 | 389 | Crystalstructure of alpha-galactosidase I from Mortierella vinacea [Umbelopsis vinacea] |
| 4NZJ_A | 6.89e-97 | 24 | 340 | 94 | 425 | Crystalstructure of a putative alpha-galactosidase (BF1418) from Bacteroides fragilis NCTC 9343 at 1.57 A resolution [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B3PGJ1 | 3.15e-183 | 20 | 400 | 23 | 402 | Alpha-galactosidase A OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=agaA PE=1 SV=1 |
| Q8VXZ7 | 1.02e-114 | 16 | 400 | 57 | 428 | Alpha-galactosidase 3 OS=Arabidopsis thaliana OX=3702 GN=AGAL3 PE=1 SV=1 |
| Q8RX86 | 1.43e-112 | 24 | 403 | 34 | 395 | Alpha-galactosidase 2 OS=Arabidopsis thaliana OX=3702 GN=AGAL2 PE=1 SV=1 |
| P14749 | 1.06e-110 | 24 | 400 | 50 | 408 | Alpha-galactosidase OS=Cyamopsis tetragonoloba OX=3832 PE=1 SV=1 |
| Q9FXT4 | 2.05e-109 | 23 | 402 | 56 | 417 | Alpha-galactosidase OS=Oryza sativa subsp. japonica OX=39947 GN=Os10g0493600 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
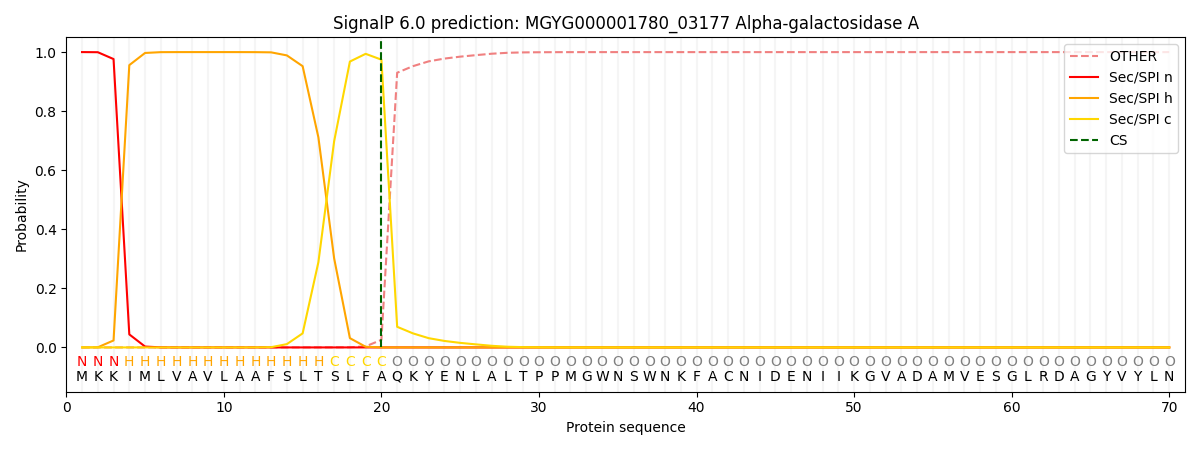
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000267 | 0.999035 | 0.000201 | 0.000170 | 0.000163 | 0.000151 |
