You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001780_04230
You are here: Home > Sequence: MGYG000001780_04230
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; | |||||||||||
| CAZyme ID | MGYG000001780_04230 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34089; End: 35153 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 52 | 311 | 2.7e-97 | 0.9963636363636363 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3934 | COG3934 | 3.33e-04 | 44 | 251 | 7 | 208 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| COG3250 | LacZ | 0.001 | 44 | 195 | 304 | 407 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 0.003 | 69 | 286 | 38 | 216 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| COG3693 | XynA | 0.007 | 146 | 220 | 105 | 185 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO68469.1 | 7.15e-255 | 1 | 353 | 1 | 353 |
| ATP57374.1 | 2.49e-149 | 27 | 349 | 28 | 349 |
| QEC78722.1 | 1.06e-145 | 12 | 349 | 9 | 347 |
| QEM13391.1 | 1.70e-142 | 5 | 349 | 8 | 348 |
| QNL49203.1 | 5.18e-140 | 25 | 349 | 43 | 366 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
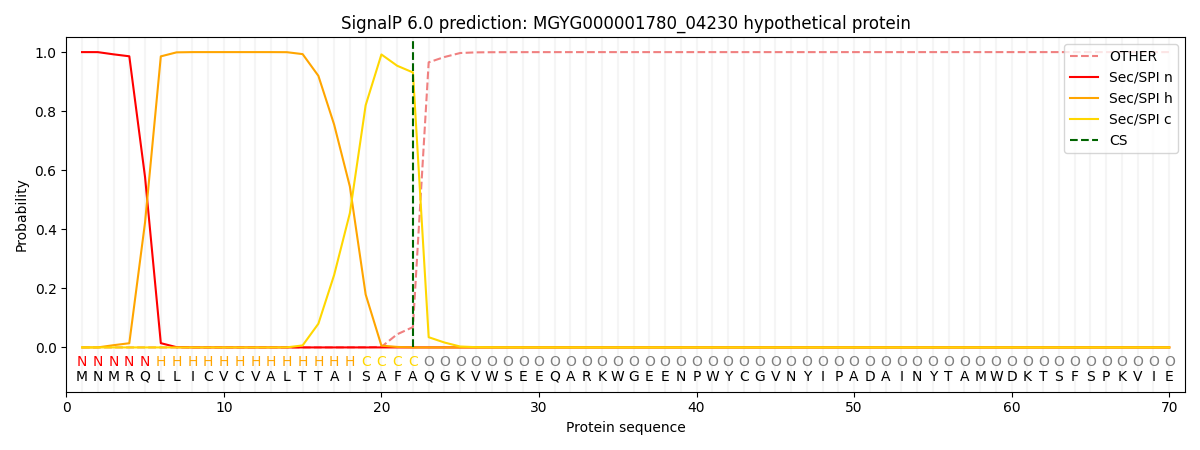
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000232 | 0.999117 | 0.000156 | 0.000170 | 0.000152 | 0.000136 |
