You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001789_01790
You are here: Home > Sequence: MGYG000001789_01790
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp002161565 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp002161565 | |||||||||||
| CAZyme ID | MGYG000001789_01790 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5506; End: 6609 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 17 | 353 | 1.8e-43 | 0.39228723404255317 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 4.12e-11 | 66 | 346 | 59 | 348 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam00703 | Glyco_hydro_2 | 1.47e-07 | 177 | 282 | 2 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY36269.1 | 6.61e-194 | 1 | 355 | 1 | 355 |
| ALJ61552.1 | 2.96e-148 | 22 | 355 | 28 | 361 |
| QUT92878.1 | 1.82e-146 | 22 | 355 | 28 | 361 |
| QBJ17536.1 | 1.36e-142 | 5 | 355 | 4 | 359 |
| QUT34134.1 | 5.38e-142 | 5 | 355 | 4 | 359 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q32JB6 | 6.06e-06 | 180 | 342 | 244 | 393 | Beta-galactosidase OS=Shigella dysenteriae serotype 1 (strain Sd197) OX=300267 GN=lacZ PE=3 SV=2 |
| B5Z2P7 | 8.02e-06 | 180 | 342 | 244 | 393 | Beta-galactosidase OS=Escherichia coli O157:H7 (strain EC4115 / EHEC) OX=444450 GN=lacZ PE=3 SV=1 |
| Q8X685 | 8.02e-06 | 180 | 342 | 244 | 393 | Beta-galactosidase OS=Escherichia coli O157:H7 OX=83334 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
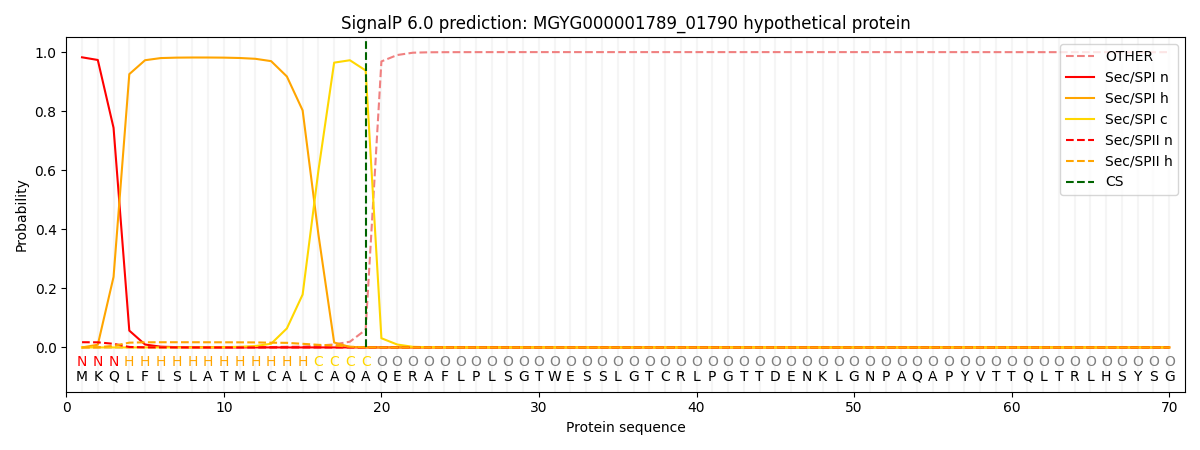
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000551 | 0.979142 | 0.019596 | 0.000278 | 0.000215 | 0.000205 |
