You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001789_02877
You are here: Home > Sequence: MGYG000001789_02877
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp002161565 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp002161565 | |||||||||||
| CAZyme ID | MGYG000001789_02877 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 193; End: 1803 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 135 | 492 | 1.6e-109 | 0.9766081871345029 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5520 | XynC | 8.76e-60 | 129 | 529 | 31 | 423 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam02055 | Glyco_hydro_30 | 3.39e-06 | 145 | 330 | 1 | 237 | Glycosyl hydrolase family 30 TIM-barrel domain. |
| cd14948 | BACON | 3.79e-04 | 41 | 127 | 7 | 83 | Bacteroidetes-Associated Carbohydrate-binding (putative) Often N-terminal (BACON) domain. The BACON domain is found in diverse domain architectures and accociated with a wide variety of domains, including carbohydrate-active enzymes and proteases. It was named for its suggested function of carbohydrate binding; the latter was inferred from domain architectures, sequence conservation, and phyletic distribution. However, recent experimental data suggest that its primary function in Bacteroides ovatus endo-xyloglucanase BoGH5A is to distance the catalytic module from the cell surface and confer additional mobility to the catalytic domain for attack of the polysaccharide. No evidence for a direct role in carbohydrate binding could be found in that case. The large majority of BACON domains are found in Bacteroidetes. |
| pfam17189 | Glyco_hydro_30C | 6.59e-04 | 449 | 498 | 3 | 52 | Glycosyl hydrolase family 30 beta sandwich domain. |
| pfam13004 | BACON | 7.81e-04 | 76 | 127 | 14 | 61 | Putative binding domain, N-terminal. The BACON (Bacteroidetes-Associated Carbohydrate-binding Often N-terminal) domain is an all-beta domain found in diverse architectures, principally in combination with carbohydrate-active enzymes and proteases. These architectures suggest a carbohydrate-binding function which is also supported by the nature of BACON's few conserved amino-acids. The phyletic distribution of BACON and other data tentatively suggest that it may frequently function to bind mucin. Further work with the characterized structure of a member of glycoside hydrolase family 5 enzyme, Structure 3ZMR, has found no evidence for carbohydrate-binding for this domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ58222.1 | 1.59e-236 | 1 | 536 | 1 | 546 |
| QUT90760.1 | 3.20e-236 | 1 | 536 | 1 | 546 |
| EDO10799.1 | 9.72e-231 | 1 | 534 | 1 | 545 |
| QRQ55177.1 | 9.72e-231 | 1 | 534 | 1 | 545 |
| ALJ48333.1 | 9.72e-231 | 1 | 534 | 1 | 545 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GTN_A | 4.40e-51 | 134 | 482 | 5 | 334 | CrystalStructure of XynC from Bacillus subtilis 168 [Bacillus subtilis],3GTN_B Crystal Structure of XynC from Bacillus subtilis 168 [Bacillus subtilis],3KL0_A Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_B Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_C Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_D Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL3_A Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_B Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_C Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_D Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL5_A Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_B Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_C Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_D Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis] |
| 1NOF_A | 1.52e-49 | 136 | 509 | 5 | 355 | ChainA, xylanase [Dickeya chrysanthemi],2Y24_A Chain A, XYLANASE [Dickeya chrysanthemi] |
| 4QAW_A | 1.99e-48 | 136 | 529 | 6 | 383 | Structureof modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_B Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_C Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_D Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_E Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_F Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_G Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_H Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 4UQA_A | 5.34e-48 | 135 | 529 | 25 | 405 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 4CKQ_A | 2.75e-47 | 135 | 529 | 25 | 405 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQ9_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQB_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQC_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQD_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQE_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q45070 | 3.90e-50 | 134 | 482 | 36 | 365 | Glucuronoxylanase XynC OS=Bacillus subtilis (strain 168) OX=224308 GN=xynC PE=1 SV=1 |
| Q6YK37 | 2.87e-48 | 136 | 535 | 39 | 422 | Glucuronoxylanase XynC OS=Bacillus subtilis OX=1423 GN=xynC PE=3 SV=2 |
| P17439 | 1.45e-07 | 131 | 333 | 83 | 335 | Lysosomal acid glucosylceramidase OS=Mus musculus OX=10090 GN=Gba PE=1 SV=1 |
| O16580 | 3.37e-07 | 127 | 344 | 80 | 349 | Putative glucosylceramidase 1 OS=Caenorhabditis elegans OX=6239 GN=gba-1 PE=1 SV=2 |
| Q70KH2 | 4.53e-07 | 131 | 493 | 103 | 517 | Lysosomal acid glucosylceramidase OS=Sus scrofa OX=9823 GN=GBA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
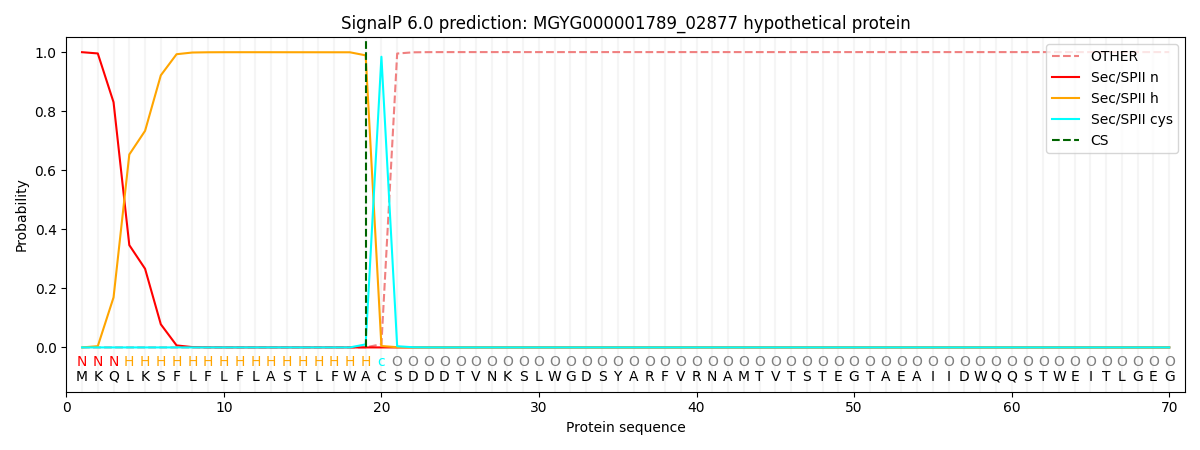
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000304 | 0.999717 | 0.000000 | 0.000000 | 0.000000 |
